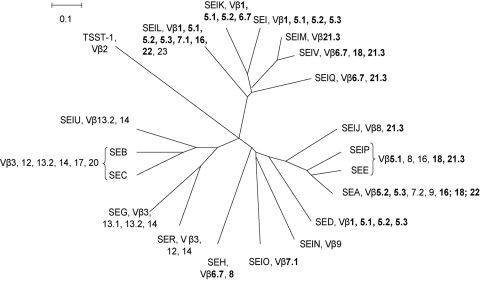
FIG. 3.
Reconstitution of the phylogenetic tree of S. aureus SAgs. Amino acid sequences of the mature toxins were deduced from sequences obtained from GenBank and SignalP analysis. Alignment was performed with ClustalX software. Evolutionary distances were determined by the Kimura method, and the values were used to construct a dendrogram by means of the neighbor-joining method using SplitsTree4 software. Vβ expansion induced by each toxin is indicated. Vβ expansions observed in response to several SSAg phylogenic groups are boldfaced.