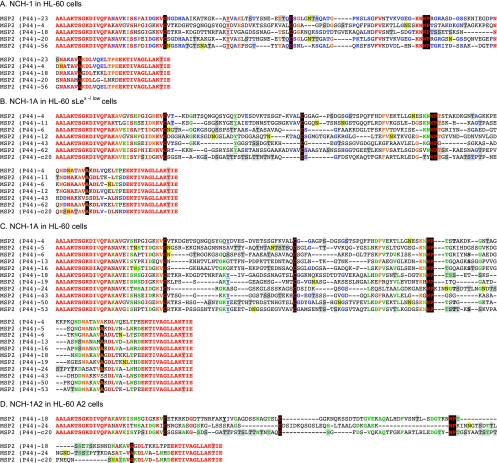
FIG. 6.
Alignment of the HVRs of Msp2(P44) paralogs expressed by NCH-1, NCH-1A, and NCH-1A2 during infection of HL-60, HL-60 sLex(−/low), and HL-60 A2 cells. The predicted amino acid sequences for the complements of msp2(p44) HVRs recovered from NCH-1 cultivated in HL-60 cells, NCH-1A cultivated in HL-60 sLex(−/low) and HL-60 cells, and NCH-1A2 cultivated in HL-60 A2 cells were aligned using Megalign. Residues that are 100% conserved in each complement of the aligned paralogs are red. Residues exhibiting 80 to 91% conservation are orange. Residues that are 64 to 73% conserved are green. Residues exhibiting 54 to 60% conservation are blue. “Signature residues” previously identified by Lin et al. (27) and Barbet et al. (2), which are very highly conserved and demarcate the most variable central portion of the HVR into three distinct segments, are highlighted with a black background. A signature tyrosine residue that may be important for intracellular replication (43) is underlined. Amino acids highlighted with a gray background are amino acids identified by NetOGlyc 3.1 (www.cbs.dtu.dk/services/NetOGlyc/) (22) and Support Vector Machines models (www.biosino.org/Oglyc) (23) as being potential O-linked glycosylation sites. Residues highlighted with a yellow background are residues predicted by NetNGlyc 1.0 (www.cbs.dtu.dk/services/NetNGlyc/) to be potential N-linked glycosylation sites.