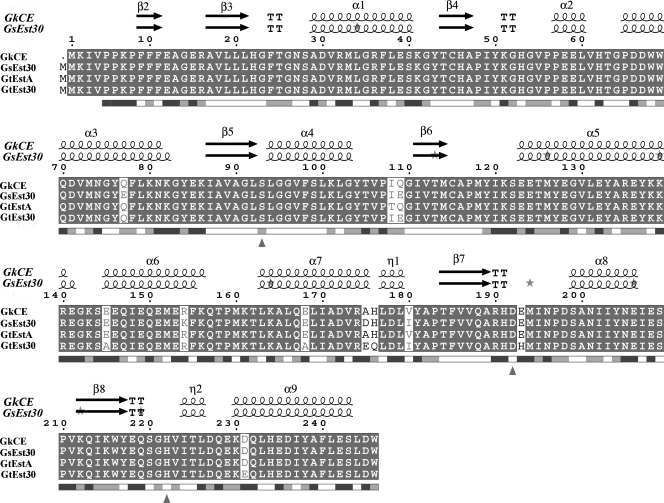
FIG. 1.
Multiple sequence alignment for CE from G. kaustophilus HTA426 (GkCE) and related Geobacillus CEs. EsPript outputs (11) obtained with the sequences from the SWISSPROT databank and aligned with CLUSTAL W (43). Sequences are grouped according to similarity. The enzyme showed 97% sequence identity with Est30Gs, 99% with EstA from G. thermoleovorans (GtEstA) NY, and 96% with Est30Gt. Residues strictly conserved have a solid background. Symbols above blocks of sequences represent the secondary structure, springs represent helices, and arrows represent β-strands. The residues forming the hydrophobic specificity pocket are indicated by small black asterisks. Triangles represent the location of the active site. K212 represents the protease cut.