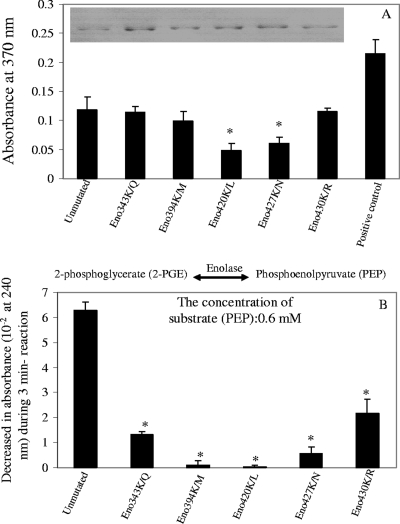
FIG. 5.
Role of lysine residues in plasminogen-binding and enzymatic activities of A. hydrophila SSU enolase. (A) Sandwich ELISA showing plasminogen-binding activities of mutated forms of Aeromonas enolase (with mutations of the indicated lysine residues). Wells of the microtiter plates were precoated with either native recombinant enolase (unmutated) or mutated forms of enolase [Eno(K343KQ), Eno(K394M), Eno(K420L), Eno(K427N), and Eno(K430R)]. An aliquot (2 μg) of plasminogen was then added to the wells for a 2-h incubation at room temperature. After a washing step, binding of plasminogen was detected by using antiplasminogen antibody. Wells coated with 2 μg of plasminogen were used as positive controls for the system while the wells coated with PBS only were used as negative controls and were set as blanks when the absorbance readings were taken. A total of eight wells were used for each group. The integrity and purity of these recombinant enolases were examined by SDS-12% PAGE with Coomassie blue staining, shown here in the inset. (B) Enzymatic activity of different forms of recombinant enolase was evaluated by measuring their ability to convert PEP to 2-PGE. Different concentrations of substrate PEP were used, and the results of a typical reaction at a 0.6 mM concentration of PEP are shown. The asterisk denotes statistically significant differences compared to the native enolase group (Student's t test).