Abstract
Escherichia coli mazEF is a toxin-antitoxin gene module that mediates cell death during exponential-phase cellular growth through either reactive oxygen species (ROS)-dependent or ROS-independent pathways. Here, we found that the stationary-phase sigma factor σS was responsible for the resistance to mazEF-mediated cell death during stationary growth phase. Deletion of rpoS, the gene encoding σS from the bacterial chromosome, permitted mazEF-mediated cell death during stationary growth phase.
Toxin-antitoxin systems have been found on the chromosomes of many bacteria (8, 10, 23, 27). One of the best studied among these chromosomal toxin-antitoxin systems is Escherichia coli mazEF, which was the first to be described as regulatable and responsible for bacterially programmed cell death (2). E. coli mazEF is located downstream from the relA gene (18, 20), specifying for ppGpp synthase (28). mazF specifies the stable toxin MazF, while mazE specifies the labile antitoxin MazE, degraded in vivo by the ATP-dependent ClpPA serine protease (2). MazF is a sequence-specific endoribonuclease that preferentially cleaves single-stranded mRNAs at ACA sequences (36, 37) and thereby inhibits translation (3, 37). MazE counteracts the action of MazF. Because MazE is a labile protein, prevention of MazF-mediated action requires the continuous production of MazE. Therefore, stressful conditions that prevent the expression of the chromosomally borne mazEF module permit the formation of free MazF and thereby cell death. These stressful conditions include (i) the use of antibiotics that are general inhibitors of transcription and/or translation such as rifampin, chloramphenicol, and spectinomycin (31); (ii) extreme amino acid starvation, leading to the production of ppGpp that inhibits mazEF transcription (2, 7); and (iii) DNA damage caused by thymine starvation (32) as well as by DNA-damaging agents like mitomycin C or nalidixic acid (11). The use of these antibiotics and other stressful conditions are well known to cause bacterial cell death (1, 5); we found that such cell death takes place through the action of the mazEF module (31, 32). All the groups of stressful conditions were found to trigger mazEF-mediated cell death by preventing the continuous synthesis of MazE and thereby reducing its level (2, 31, 32). We were surprised to find that mazEF-mediated cell death occurs at the exponential stage of growth but does not occur during stationary phase (11).
We have recently reported that the activation of E. coli mazEF by using stressful conditions causes the generation of reactive oxygen species (ROS) (15). ROS have been previously implicated in programmed cell death in eukaryotes (21, 29), including in yeast (12, 17), in the life span of several organisms (25, 33), in the senescence of bacteria (6), and in the mode of action of some antibiotics (13-15). It was previously reported that the stationary-phase sigma factor σS, encoded by rpoS (16, 19), positively regulates the formation of catalase and is responsible for the elevated levels of this enzyme during stationary growth phase (24, 34, 35). Since catalase detoxifies ROS, we asked whether resistance of stationary-phase cells to mazEF-mediated cell death was caused by the elevated levels of catalase produced at that time. So, we tested whether deleting rpoS from E. coli cells would lead to their death through the mazEF system during stationary growth phase. Indeed, as we have predicted, in ΔrpoS cells we observed mazEF-mediated cell death even during stationary phase of growth.
We used strain MC410relA+ and its ΔmazEF::kan derivative (9) and strain MC4100relA+ΔrpoS, which we constructed by P1 transduction from E. coli strain K-38ΔrpoS::tet (kindly provided by Shosh Altuvia), and its ΔmazEF derivative, which we constructed by PCR deletion (4). We used plasmid pQEkatE (15), bearing the catalase-specifying katE gene, which is continuously expressed in the strains described here.
We grew the bacteria in liquid M9 minimal medium with 1% glucose and a mixture of amino acids (10 μg/ml each) (22) and then plated them on rich LB agar plates, as we have described previously (11).
Nalidixic acid, mitomycin C, trimethoprim, rifampin, serine hydroxamate, chloramphenicol, spectinomycin, Trizma base, sodium dodecyl sulfate, DNase, and RNase were obtained from Sigma (St. Louis, MO). Lysozyme was obtained from the United States Biochemical Corporation (Cleveland, OH). Ampicillin was obtained from Biochemie GmbH (Kundl, Austria). Carbonylated proteins were detected using the chemical and immunological reagents from the OxyBlot oxidized protein detection kit (Chemicon, Temecula, CA). Nitrocellulose membranes were obtained from Pall Corporation (New York). Luminol and p-coumaric acid were obtained from Sigma (St. Louis, MO), hydrogen peroxidase solution was obtained from Merck (NJ), and AnaeroGen bags were obtained from Gamidor Diagnostics (Petach Tikva, Israel).
We studied the effects of using various stressful conditions on cell viability during stationary phase under aerobic conditions as follows. We diluted (1/100) an overnight culture in M9 medium and grew the cells while shaking them (160 rpm) in the same medium at 37°C either until they reached exponential growth phase (optical density at 600 nm [OD600], 0.6) or until they reached stationary growth phase (for about 16 h; OD600, about 1.6). When the cultures reached either exponential or stationary growth phase, we incubated aliquots of the cells at 37°C for 10 min without shaking them. We then submitted the cell aliquots to various stressful conditions (described in the legends to the figures), plated the cells on LB agar, and incubated them at 37°C overnight. For each strain, we determined the ratio of CFU of treated cells versus that of untreated cells.
We studied the effects of using various stressful conditions on cell viability during stationary phase under anaerobic conditions as follows. We grew the cells in 15-ml tubes containing 10 ml of M9 medium while standing and without shaking them in an anaerobic jar containing AnaeroGen bags at 37°C. We incubated the cells for 3 to 4 days until the cultures reached an OD600 of 1.4. We then transferred 1-ml samples to 1.5-ml Eppendorf tubes and incubated them further in the anaerobic jar at 37°C for 10 min. After incubating the cells, we induced stressful conditions under anaerobic conditions, as described in a figure legend (see Fig. 3). The cells were centrifuged, washed, diluted, plated, and incubated in the anaerobic jar at 37°C for 20 h.
FIG. 3.
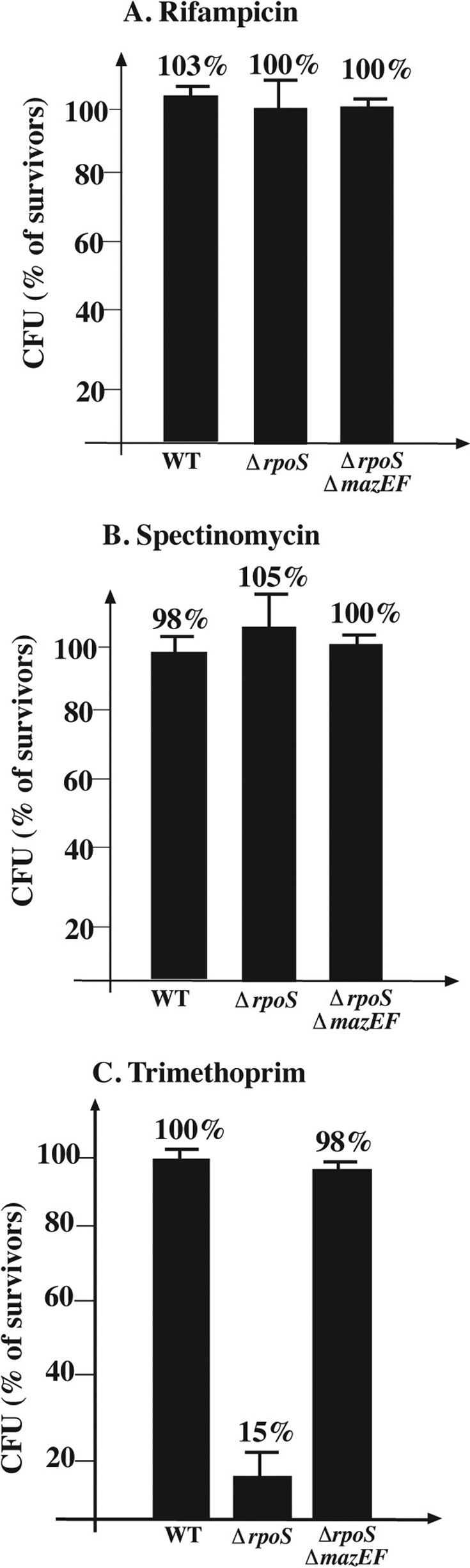
The effect of E. coli rpoS on mazEF-mediated cell death during stationary phase under anaerobic conditions. Strains described in the legend of Fig. 1 were grown under anaerobic conditions until the stationary phase (OD600 = 1.3 to 1.4). Stressful conditions were induced by incubation of cells at 37°C, without shaking them, with rifampin (10 μg/ml) for 10 min (A); spectinomycin (1 mg/ml) for 10 min (B); or trimethoprim (5 μg/ml) for 1 h (C). For the rest of the experimental details, see the text.
Deletion of rpoS permitted E. coli mazEF-mediated cell death during stationary growth phase.
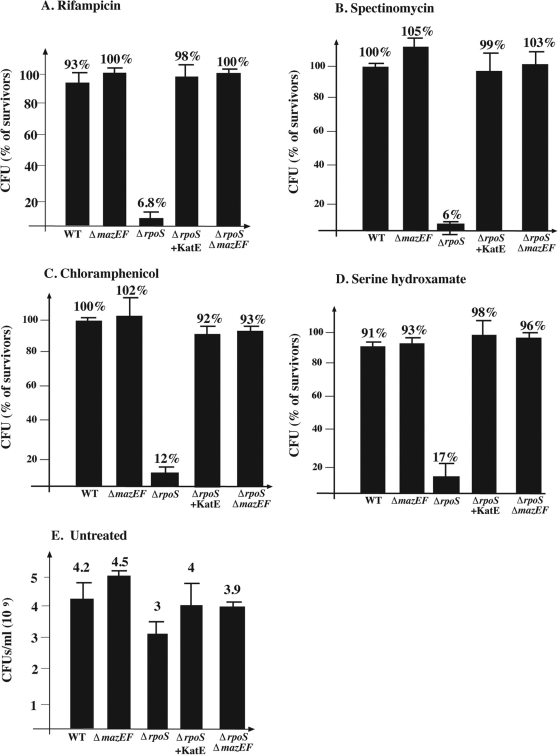
We have reported previously that E. coli mazEF-mediated cell death occurs during exponential phase but not during stationary phase (11). Here, we studied the role of the stationary-phase sigma factor σS, encoded by rpoS (16, 19, 30), for two reasons as follows. (i) σS positively regulates the formation of catalase, an ROS-detoxifying enzyme, and is responsible for the elevated levels of this enzyme during stationary phase. (ii) We found previously (15) that catalase prevents mazEF-mediated cell death induced by inhibitors of transcription or translation. So, we tested whether deleting rpoS from E. coli cells under various stressful conditions would lead to their death through the mazEF system during stationary growth phase. We found that during stationary phase, though the wild-type (WT) cells did not die, we did observe mazEF-mediated cell death in the ΔrpoS cells (Fig. 1 and 2). Note that in untreated stationary-phase cultures, deleting rpoS caused a reduction in cell viability of only 30% (Fig. 1E).
FIG. 1.
The effect of E. coli rpoS on mazEF-mediated cell death during stationary growth phase following the inhibition of transcription or translation. MC4100relA+ (WT) and its derivatives, MC4100relA+ΔmazEF::kan (ΔmazEF), MC4100relA+ΔrpoS::tet (ΔrpoS), MC4100relA+ΔrpoS::tet carrying plasmid pQEkatE (ΔrpoS+KatE), and MC4100relA+ΔrpoSΔmazEF (ΔrpoS ΔmazEF) were grown aerobically at 37°C until stationary phase (OD600 = 1.3 to 1.4). Stressful conditions were induced by incubation of cells at 37°C, without shaking them, with rifampin (20 μg/ml) for 10 min (A); spectinomycin (1 mg/ml) for 10 min (B); chloramphenicol (50 μg/ml) for 20 min (C); or serine hydroxamate (0.2 mg/ml) for 1 h (D); for the rest of the experimental details, see the text. The results represent the ratio of CFU of treated cells versus that of untreated cells. (E) CFU/ml of all untreated strains at stationary phase.
FIG. 2.
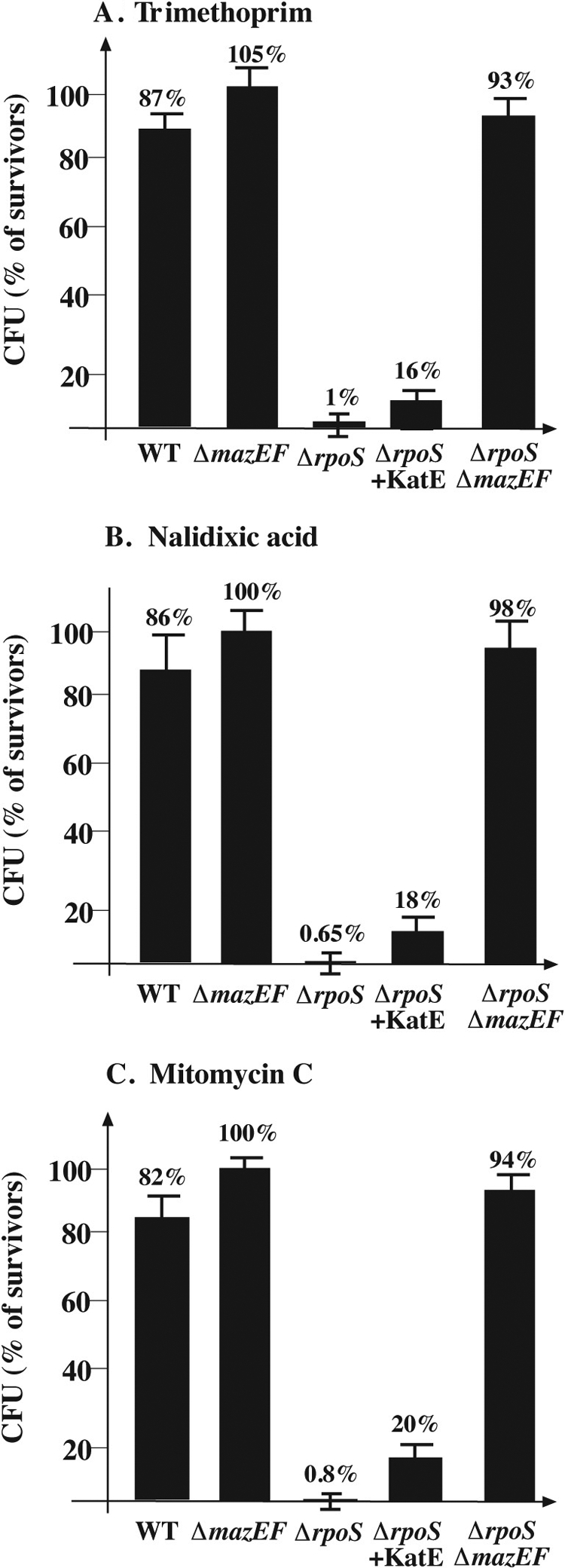
The effect of E. coli RpoS on mazEF-mediated cell death during stationary phase following DNA damage. Strains described in the legend to Fig. 1 were grown aerobically at 37°C until stationary phase (OD600 = 1.3 to 1.4). Stressful conditions were induced by incubation of cells at 37°C, without shaking them, with trimethoprim (5 μg/ml) for 1 h (A); nalidixic acid (1 mg/ml) for 10 min (B); or mitomycin C (0.25 μg/ml) for 10 min (C). For the rest of the experimental details, see the text.
mazEF-mediated cell death operates through an ROS-dependent and ROS-independent pathway in an ΔrpoS mutant at stationary phase.
We found that during stationary growth, mazEF-mediated cell death induced by inhibitors of transcription and translation occurred in ΔrpoS cells and was also prevented by the overproduction of catalase (Fig. 1). On the other hand, mazEF-mediated cell death induced by DNA-damaging agents like trimethoprim, nalidixic acid, and mitomycin C was not prevented by the overproduction of catalase (Fig. 2). This additional result supports our previous finding that DNA-damaging agents induce mazEF-mediated cell death through an ROS-independent pathway (15). Note, however, that even though overproducing catalase did not completely prevent cell death induced by DNA damage in an ΔrpoS strain during stationary growth, it improved cell viability by about 18 times (from 1% to 18%) (Fig. 2).
Obviously, ROS are not formed in the absence of oxygen. We studied the effect of using completely anaerobic conditions on mazEF-mediated cell death during stationary growth. We activated mazEF by adding rifampin to inhibit transcription (Fig. 3A), by adding spectinomycin to inhibit translation (Fig. 3B), or by adding trimethoprim to cause DNA damage (Fig. 3C). We observed no mazEF-mediated cell death in an ΔrpoS strain grown anaerobically when we added antibiotics that inhibited transcription (rifampin) or translation (spectinomycin) (Fig. 3A). However, in an ΔrpoS mutant, when the mazEF module was activated by the DNA-damaging agent trimethoprim, we observed mazEF-mediated cell death even under anaerobic growth conditions (Fig. 3C). This suggests that in the ΔrpoS mutant, mazEF-mediated cell death induced by DNA damage was ROS independent. Our results obtained under anaerobic conditions (Fig. 3) confirmed those obtained with catalase (Fig. 1 and 2).
mazEF-mediated protein carbonylation at stationary phase.
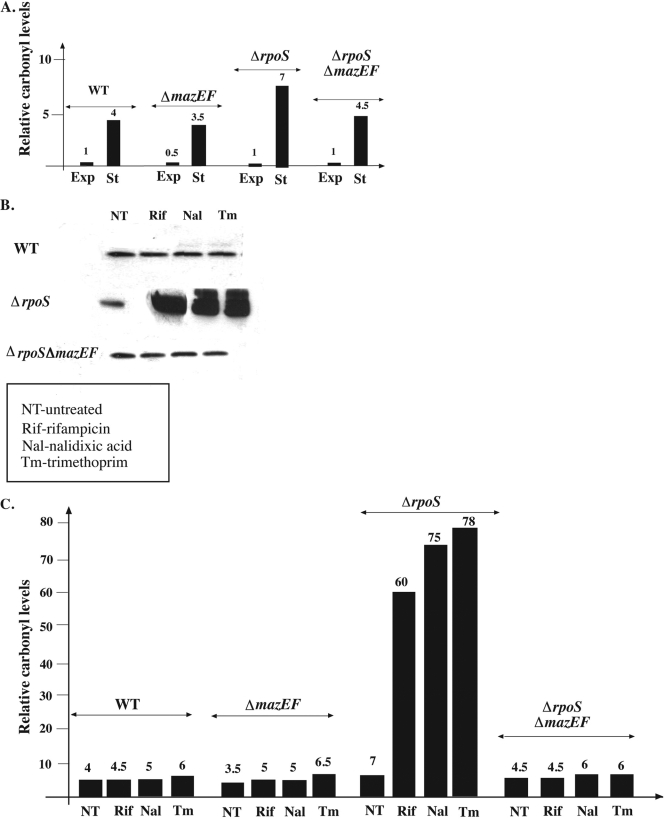
Carbonylated proteins are oxidized proteins that carry carbonyl groups generated by ROS (6). Previously, it was shown that elevated levels of protein carbonylation are formed during stationary growth phase (6). Here, we also observed that, in WT cells, protein carbonylation was elevated four times more during stationary growth phase than during exponential growth phase (Fig. 4A). Since we observed similar results with the ΔmazEF mutant (Fig. 4A), it seems that the increase in protein carbonylation was mazEF independent. We have previously described mazEF-dependent protein carbonylation under stressful conditions (15). Such an effect was not observed here in the stationary-phase cultures of the WT and its ΔmazEF mutant (Fig. 4B and C). However, we observed a dramatic increase in the level of carbonylation in the ΔrpoS mutant compared with that of the WT strain at stationary phase; applying stressful conditions (rifampin, nalidixic acid, or trimethoprim) increased protein carbonylation by about 10 times in the ΔrpoS mutant (Fig. 4C). Since we did not observe this dramatic increase in protein carbonylation in the ΔrpoS ΔmazEF double mutant, this phenomenon must be mazEF dependent.
FIG. 4.
mazEF-mediated carbonylation of cellular protein during stationary growth following the induction of mazEF by various stressful conditions. MC4100relA+ (WT) and its derivatives MC4100relA+ΔmazEF (ΔmazEF), MC4100relA+ΔrpoS::tet (ΔrpoS), and MC4100relA+ΔrpoSΔmazEF (ΔrpoS ΔmazEF) were grown to either exponential or stationary phase. Stressful conditions were induced as described in the legends to Fig. 1 and 2. Cells that were either not treated or treated under stressful conditions were lysed as we have described previously (15). In these lysates, we examined the level of protein carbonylation using the Chemicon OxyBlot kit to derivatize the carbonyl groups in the protein side chains to 2,4-dinitrophenylhydrazone (DNP-hydrazone) by reaction with 2,4-dinitrophenylhydrazine. The DNP-derivative crude proteins were detected using a primary antibody specific to the DNP moiety of the proteins (for details, see reference 15). (A) Relative protein carbonylation of untreated exponential-phase cultures (Exp) and untreated stationary-phase cultures (St). Protein carbonylation was determined and quantified, as we have described previously (15). The first column represents the carbonyl level of untreated exponential-phase WT cells, which was arbitrarily determined to be 1. The subsequent columns represent the relative carbonyl level of each strain compared with the levels of the exponential-phase untreated WT cells. (B) Protein carbonylation of stationary-phase cultures submitted to various stressful conditions. Carbonylated proteins were detected. (C) Relative carbonyl levels presented in panel B. The intensity of each band presented in panel B was quantified, as we have described previously (15). The columns represent the relative carbonyl level of each of the treated strains compared with the level in untreated, exponentially growing WT cells (first column in panel A).
A new finding for this study is that E. coli mazEF-mediated cell death can occur during the stationary phase of growth. We found that at this stage of growth, σS, the stationary-phase sigma factor encoded by rpoS, is a key component. We observed mazEF-mediated cell death triggered by various stressful conditions (Fig. 1 and 2) in an ΔrpoS strain but not in the isogenic WT strain. Based on these results, we hypothesize that rpoS may antagonize mazEF-mediated cell death with at least two mechanisms as follows. (i) The induction of the katE gene (24, 34), which inhibits ROS formation, is one of these mechanisms (30). This hypothesis is supported by our results, showing that either the overproduction of catalase or use of completely anaerobic conditions complements the effect of rpoS deletion, thus leading to the prevention of mazEF-mediated cell death (Fig. 1). (ii) When mazEF is triggered by DNA damage, RpoS may antagonize mazEF-mediated cell death by a different mechanism than by induction of the katE gene. In this case, the overproduction of catalase or use of completely anaerobic conditions only slightly improved cell survival (Fig. 2). It is well known that the rpoS gene product σS is a global regulatory protein associated with stationary growth of bacterial cultures (16, 26, 34). We hypothesize that at least one of the gene products controlled by σS may antagonize the putative death executioner protein(s) of the ROS-independent mazEF death pathway.
Thus, mazEF-mediated cell death is a programmed phenomenon that is stress induced and normally takes place only during exponential growth phase. It does not take place during stationary growth phase, not because the death program is missing but rather because it is antagonized by ROS-detoxifying enzymes and by another as yet unidentified cellular component(s).
Acknowledgments
We thank F. R. Warshaw-Dadon (Jerusalem, Israel) for her critical reading of the manuscript.
This research was supported by grant 177/07 from the Israel Science Foundation (ISF), administrated by the Israel Academy of Sciences and Humanities, by grant 2005029 from the United States-Israel Binational Science Foundation (BSF), and by grant GM069509 from the National Institutes of Health.
Footnotes
Published ahead of print on 27 February 2009.
REFERENCES
- 1.Ahmad, S. I., S. H. Kirk, and A. Eisenstark. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 52591-625. [DOI] [PubMed] [Google Scholar]
- 2.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 936059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332809-819. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, W. 1998. Antibiotic resistance in bacteria, p. 239-273. In R. M. Krause (ed.), Emerging infections. Academic Press, New York, NY.
- 6.Dukan, S., and T. Nystrom. 1998. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 123431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 5343-70. [DOI] [PubMed] [Google Scholar]
- 8.Engelberg-Kulka, H., R. Hazan, and S. Amitai. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 1184327-4332. [DOI] [PubMed] [Google Scholar]
- 9.Engelberg-Kulka, H., M. Reches, S. Narasimhan, R. Schoulaker-Schwarz, Y. Klemes, E. Aizenman, and G. Glaser. 1998. rexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci. USA 9515481-15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelberg-Kulka, H., B. Sat, M. Reches, S. Amitai, and R. Hazan. 2004. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 1266-71. [DOI] [PubMed] [Google Scholar]
- 11.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 1863663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herker, E., H. Jungwirth, K. A. Lehmann, C. Maldener, K. U. Frohlich, S. Wissing, S. Buttner, M. Fehr, S. Sigrist, and F. Madeo. 2004. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130797-810. [DOI] [PubMed] [Google Scholar]
- 14.Kohanski, M. A., D. J. Dwyer, J. Wierzbowski, G. Cottarel, and J. J. Collins. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolodkin-Gal, I., B. Sat, A. Keshet, and H. Engelberg-Kulka. 2008. The communication factor EDF and the toxin-antitoxin module mazEF determine the mode of action of antibiotics. PLoS Biol. 6e319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 549-59. [DOI] [PubMed] [Google Scholar]
- 17.Madeo, F., S. Engelhardt, E. Herker, N. Lehmann, C. Maldener, A. Proksch, S. Wissing, and K. U. Frohlich. 2002. Apoptosis in yeast: a new model system with applications in cell biology and medicine. Curr. Genet. 41208-216. [DOI] [PubMed] [Google Scholar]
- 18.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 1756850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCann, M. P., J. P. Kidwell, and A. Matin. 1991. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 1734188-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metzger, S., I. B. Dror, E. Aizenman, G. Schreiber, M. Toone, J. D. Friesen, M. Cashel, and G. Glaser. 1988. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J. Biol. Chem. 26315699-15704. [PubMed] [Google Scholar]
- 21.Mignotte, B., and J. L. Vayssiere. 1998. Mitochondria and apoptosis. Eur. J. Biochem. 2521-15. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Mittenhuber, G. 1999. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biotechnol. 1295-302. [PubMed] [Google Scholar]
- 24.Mulvey, M. R., J. Switala, A. Borys, and P. C. Loewen. 1990. Regulation of transcription of katE and katF in Escherichia coli. J. Bacteriol. 1726713-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami, S., and T. E. Johnson. 1998. Life extension and stress resistance in Caenorhabditis elegans modulated by the tkr-1 gene. Curr. Biol. 81091-1094. [DOI] [PubMed] [Google Scholar]
- 26.Nystrom, T. 2004. Stationary-phase physiology. Annu. Rev. Microbiol. 58161-181. [DOI] [PubMed] [Google Scholar]
- 27.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potrykus, K., and M. Cashel. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 6235-51. [DOI] [PubMed] [Google Scholar]
- 29.Raha, S., and B. H. Robinson. 2001. Mitochondria, oxygen free radicals, and apoptosis. Am. J. Med. Genet. 10662-70. [DOI] [PubMed] [Google Scholar]
- 30.Sak, B. D., A. Eisenstark, and D. Touati. 1989. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc. Natl. Acad. Sci. USA 863271-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 1832041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 1851803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sohal, R. S., S. Agarwal, A. Dubey, and W. C. Orr. 1993. Protein oxidative damage is associated with life expectancy of houseflies. Proc. Natl. Acad. Sci. USA 907255-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijayakumar, S. R., M. G. Kirchhof, C. L. Patten, and H. E. Schellhorn. 2004. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J. Bacteriol. 1868499-8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Ossowski, I., M. R. Mulvey, P. A. Leco, A. Borys, and P. C. Loewen. 1991. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J. Bacteriol. 173514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Y., J. Zhang, H. Hara, I. Kato, and M. Inouye. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2803143-3150. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12913-923. [DOI] [PubMed] [Google Scholar]