Abstract
Mycoplasma genitalium is a human bacterial pathogen linked to urethritis and other sexually transmitted diseases. Here, we assessed the incidence of M. genitalium infection in patients attending a sexually transmitted disease clinic in San Antonio, TX, by use of diagnostic real-time PCR. Overall, 16.8% of women and 15.1% of men were found M. genitalium positive. Regions of the mgpB gene, which encodes the MgPa adhesin, were amplified from positive clinical specimens and evaluated for sequence variability, which demonstrated transmission of the pathogen between sexual partners. Follow-up analysis of a subset of patient specimens revealed reinfection by a different strain of M. genitalium, indicating the absence of protective immunity. Eighteen DNA sequence variants were obtained and compared with all other available clinical sequences. Detailed analysis revealed silent mutations of six amino acid residues within the encoded region of the MgPa adhesin in numerous clinical strains. In addition, missense mutations of limited numbers of amino acids were observed. Alignment of putative amino acid sequences revealed the simultaneous occurrence of several mutations and the existence of identical or similar protein variants in strains from different locations.
Mycoplasma genitalium is associated with numerous human genitourinary tract maladies, including nonchlamydial, nongonococcal urethritis in men and cervicitis, endometritis, and pelvic inflammatory disease in women (1, 4, 5, 12, 19, 29, 30, 35, 39). Understanding the epidemiological aspects of infections and developing effective treatment require reliable typing of pathogenic microorganisms. Since M. genitalium is very difficult to culture from clinical specimens (13, 21), classical microbiological typing methods are not readily applicable. Furthermore, serological approaches are not widely used because of cross-reactivity with other mycoplasmas, especially with Mycoplasma pneumoniae (15, 26). Therefore, typing of M. genitalium strains relies on DNA sequence data. Recently, sequence variability of the rRNA operon and tandem repeats in the MG309 locus have been evaluated with clinical specimens (28). However, the majority of M. genitalium clinical sequence data available today is based upon the gene mgpB (locus MG191 of sequenced reference strain G37 [11]) encoding the M. genitalium adhesin MgPa (14, 17, 21). An M. genitalium-specific diagnostic PCR assay was designed to target the proximal unique region of this gene (using primers MgPa-1 and MgPa-3 [22] and designated here “MgPa-13”). The MgPa-13 region has exhibited sequence stability in persistently infected patients for up to 21 months (17). Despite this intrastrain stability, high levels of sequence variability between clinical isolates were observed (14, 17). Based on this sequence region, transmission of M. genitalium between sexual partners was shown, as was colonization of different anatomical sites of the same patient by identical strains (14).
Here, we present analysis of MgPa-13 sequences generated from clinical specimens collected at the Project S.A.F.E. sexually transmitted disease (STD) clinic in San Antonio, TX (36), from recruited female participants and their male partners. Alignment of MgPa-13 sequences corroborated transmission between partners and colonization of different anatomical sites by the same strain. Comparison of newly recovered sequences with all currently available clinical data (14, 17, 21) revealed not only the presence of several common variants but also distinct sequences. Analyses of encoded amino acids uncovered mutations that determine common features among MgPa-13 region variants.
MATERIALS AND METHODS
Origin and processing of clinical specimens.
A group of minority women (Mexican and African American) and their partners were recruited from public health clinics throughout San Antonio, TX, into a controlled randomized trial of behavioral-cognitive interventions to reduce the recurrence of STDs (36). Women were recruited into the study after being diagnosed with a nonviral STD, specifically infection with Chlamydia trachomatis, Neisseria gonorrheae, Trichomonas vaginalis, or Treponema pallidum. At each clinic visit, urine specimens were obtained from both female and male subjects, and at the same time, vaginal and cervical swab (VC) specimens were collected from female participants. All urine and VC samples were transported to the University of Texas Health Science Center at San Antonio (UTHSCSA) and were processed immediately using centrifugation and resuspension of pellets in water prior to boiling (2). Specimens examined here were obtained from 500 patient visits during a 13-month period (September 2005 to October 2006).
Detection of M. genitalium.
Detection of M. genitalium was performed by quantitative duplex real-time PCR assay with inhibition control (20). Serial dilutions representing from 1 to 108 genome copies of M. genitalium reference strain G37 (14th passage) were used to determine the efficiency and detection limit of the assay. We consistently detected five or more genomes per reaction and one genome per reaction with 60% probability. Five dilutions ranging from 1 to 107 genomes were included as standard concentrations on each test plate for quantification. Duplex TaqMan assays were performed on 5-μl aliquots of DNA preparation in 50-μl reaction mixtures with universal master mix, as suggested by the manufacturer (Applied Biosystems). Individual aliquots represented 400 μl of urine specimens or ∼1% of collected VC specimens. Assays were performed using the ABI Prism 7900HT sequence detection system.
PCR amplifications of MgPa-13 regions.
With M. genitalium-positive specimens, amplification of MgPa-13 regions was performed using primers MgPa-1 (5′-AGTTGATGAAACCTTAACCCCTTGG-3′) and MgPa-3 (5′-CCGTTGAGGGGTTTTCCATTTTTGC-3′) (22) and Platinum Taq DNA high-fidelity polymerase system, as suggested by the manufacturer (Invitrogen). Chromosomal DNA isolated from reference strain G37 (100 ng) was used as a positive control alongside a control for contamination of reagents. All amplified products were visualized on 2% agarose and purified (QIAquick PCR purification kit; Qiagen) prior to sequencing.
Sequence analysis.
Sequencing of both strands from M. genitalium-positive specimens was accomplished by the Department of Microbiology and Immunology Nucleic Acids Facility, UTHSCSA. DNA and amino acid sequence alignments and phylogenetic tree predictions were performed by CLUSTALW (Fig. 1). The sequence of the corresponding region of the M. pneumoniae adhesin P1 gene (MPN141) was included as an outlier. Positions of amino acid residues (Fig. 2 and 3) correspond to the protein sequence in G37.
FIG. 1.
Predicted phylogenetic tree. Reference strain G37 and 73 distinct clinical sequences obtained in this study or previously (14, 17, 21) were aligned by CLUSTALW. The sequence of the corresponding region of the M. pneumoniae adhesin P1 gene (MPN141) was included as an outlier.
FIG. 2.
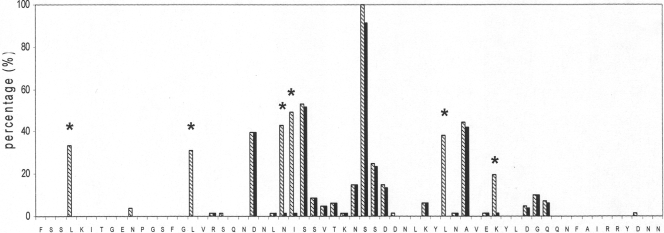
Comparison of MgPa-13 nucleotide and deduced amino acid sequences. The region of F75 (extreme left) to N136 (extreme right) of the MgPa protein in reference strain G37 is presented. Percentages of mutated codons (striped bars) and of detected missense mutations (black bars) are shown. Percentages represent the fraction of the 73 aligned clinical sequences that contained a codon/amino acid different from that of the G37 sequence. Recurrent silent mutations of six amino acid residues are indicated (asterisks).
FIG. 3.
Alignment of deduced amino acid sequence from F75 to N136 of reference strain G37-MgPa with sequences of clinical strains. Amino acids identical to those of G37 are presented as dots (•); missense mutations are highlighted with gray (hydrophobic amino acid) or black (hydrophilic amino acid). Amino acid deletions compared with those of G37-MgPa are indicated (−). Positions of the majority of silent mutations are indicated (asterisks). Sequences were grouped to demonstrate existence of observed protein variants. Seventy-three distinct clinical sequence variants were obtained previously (14, 17, 21) and in this study.
Sequence designation.
Sequences are identified by numbers assigned to the specimen upon arrival to the laboratory. If identical sequences were generated from both partners, only the sequence from the woman is presented. If identical sequences were obtained from unrelated participants (i.e., different participants with no known sexual relationship), only the sequence with the earliest collection date is presented.
Nomenclature.
The major M. genitalium adhesin MgPa (16) (also known as protein P140 [33]) is encoded by locus MG191 of the sequenced reference strain G37 (gene mgpB, nucleotides 221570 to 225904). In this paper we refer to the analyzed region as MgPa-13, since it is amplified using primers MgPa-1 and MgPa-3 (22).
Nucleotide sequence accession numbers.
Sequences of MgPa-13 regions obtained for clinical specimens collected in the San Antonio, TX, area have been submitted to the GenBank database and assigned accession numbers FJ750827 to FJ750844.
RESULTS
Incidence of M. genitalium in patient population.
Among specimens collected from 268 females, 11.5% of urine specimens and 10.9% of VC specimens were positive for M. genitalium. Overall, 16.8% women were M. genitalium positive (based upon at least one of the analyzed specimens being positive) and of those, 15 were diagnosed positive by both urine and VC. Of 232 male participants, 15.1% were M. genitalium positive. Using a quantitative real-time PCR assay, the number of M. genitalium genomes per reaction in positive specimens ranged from 1 to ∼10,000.
All M. genitalium-positive specimens were used to amplify MgPa-13 regions. Limited volumes of specimens and low genome numbers of M. genitalium in many positive samples (only 30% of specimens contained 10 or more M. genitalium genomes per reaction) resulted in the generation of 31 MgPa-13 amplicons. Only one sequence variant was obtained from each specimen. Sequences generated from related specimens (i.e., same patients or sexual partners) were excluded from further examination, and DNA sequence analyses revealed 18 different sequences. The highest M. genitalium load was found in specimen TX135 during the patient's initial visit. On her second visit (after 6 months), she was M. genitalium negative. On her subsequent two visits (after 18 and 20 months), she was again M. genitalium positive and the generated MgPa-13 sequences were identical (TX1428). However, alignment with TX135 showed that she was reinfected by a different strain of M. genitalium (differences in seven nucleotides).
Sequences generated from different anatomical sites of female participants were identical.
As mentioned earlier, both urine and VC specimens collected from 15 female patients were M. genitalium positive, and we examined MgPa-13 regions from both specimens of 4 women. Comparison of these sequences showed that each woman was infected by a different M. genitalium strain. Additionally, identical sequences were obtained from both anatomical sites of the patient. This demonstrated colonization of each patient by a single M. genitalium strain, as previously observed (14).
Partners were infected by the same M. genitalium strain.
The majority of the analyzed specimens were collected from sexual partners (172 dyads). We detected M. genitalium in both partners in 19 couples (11% of participating dyads). All positive dyad specimens were further used for MgPa-13 region amplification, and we generated paired products for four dyads. Analysis of these MgPa-13 regions revealed that sequences were identical between partners, which corroborated sexual transmission of M. genitalium among partners (14).
Both common and unique MgPa-13 variants identified among Texas strains.
Comparison of DNA sequences generated from this study with all currently available clinical sequences (14, 17, 21) revealed identical sequences. For example, sequences indistinguishable from TX135, TX302, and TX1428 were found among previously published sequences (14, 17). However, 13 DNA sequence variants specific to the San Antonio area were identified.
For phylogenetic tree prediction, 73 distinct clinical sequences of the MgPa-13 region were compared with reference strain G37. Identified common sequences were represented by their earliest published types, and consequently, the San Antonio area sequences and the Kenya collection (17) were represented only by their unique sequences. Results suggested that site-specific M. genitalium strains were endemic in different areas and are presented in Fig. 1.
Alignment of putative amino acid sequences of the MgPa-13 region reveals high occurrence of silent mutations.
When putative amino acid sequences encoded by all unique clinical variants were examined, strains had uninterrupted reading frames (amino acids 61 to 153 of G37-MgPa). Upon comparison of the deduced amino acid sequences in clinical strains with the G37-MgPa sequence, a high occurrence of silent mutations of six codons was observed (Fig. 2 and Table 1). They represented 38% of all detected codon mutations. It also appears that several of the silent mutations were found together in the same sequence variant. For example, among 26 strains with the L78 silent mutation, 23 strains also carry the I100 silent mutation (out of 34); 24 strains carry the L115 silent mutation (out of 28); 21 strains carry the L90 silent mutation (out of 23); 21 strains carry the N99 silent mutation (out of 30); and 12 strains carry the K120 silent mutation (out of 15). Despite the DNA sequence variability, amino acid residues were identical.
TABLE 1.
Typical silent mutations observed with MgPa-13 region of M. genitalium clinical strainsa
| Amino acid | Codon changeb | No. of strains with mutated codon |
|---|---|---|
| L78 | CTA (1.26) → CTG (0.43) | 26 |
| L90 | TTA (5.03) → CTA (1.26) | 23 |
| L115 | CTC (0.49) → CTT (1.99) | 28 |
| N99 | AAT (4.61) → AAC (2.89) | 30 |
| I100 | ATT (5.18) → ATC (1.78) | 34 |
| K120 | AAA (7.03) → AAG (2.43) | 15 |
Missense mutations lead to distinct sequence variants in MgPa.
The majority of missense mutations were limited to a restricted number of amino acids (Fig. 2 and 3) with noticeably favored codon mutations. For example, 23 D96 mutations (out of 28 detected) were due to replacement of GAC by GAG (encoding E) and four were GAC-to-GCG (encoding A) mutations. Interestingly, simultaneous occurrence of several missense mutations was also detected with strains of different geographic origin, resulting in identical variants of the protein (Fig. 3). For example, simultaneous substitutions of D96 by E, S101 by A, and S107 by D/G were found in 20 strains. Another recognized variant contained simultaneous D96-to-A, S101-to-A, T104-to-Q, N106-to-Q, and S108-to-G mutations.
DISCUSSION
The M. genitalium mgpB gene consists of regions that share homologies with nine repetitive sequences (MgPar) spread throughout the chromosome (7, 17). Extensive sequence variability resulting from recombination between repetitive elements of mgpB and MgPars was recently confirmed with reference strain G37 as well as with clinical strains (17, 18, 27). However, the MgPa-13 region (nucleotides 181 to 460) does not share homology with other regions in the G37 genome, and this region was chosen for development of a conventional diagnostic PCR assay (22). Later, longitudinal sequence stability within a strain was demonstrated (14, 17), and the use of this region for typing purposes was proposed. Analysis of a growing number of clinical data uncovered significant interstrain sequence variability. Sequences of the MgPa-13 region represent the largest set of data generated from different clinical strains of M. genitalium (13, 14, 17).
During a 13-month period (2005 to 2006), we evaluated M. genitalium incidence with patients recruited under Project S.A.F.E. in San Antonio, TX (25, 36). Specimens (urine and VC swab) were collected during 500 visits and assessed using a diagnostic M. genitalium-specific real-time PCR assay (20). Overall, 16.8% of women and 15.1% of men were found M. genitalium positive. These numbers are higher than results of similar studies of STD clinic attendees (1, 3, 10, 29, 38), which could be explained by the facts that our specimen collection was conducted over a rather short time period and the patient population consisted of women (and their partners) who were recruited after being diagnosed with nonviral STDs (36).
The MgPa-13 region was further amplified from M. genitalium-positive specimens, and sequence analysis revealed a high degree of variability among local strains. From the originally amplified 31 sequences, we eliminated related (i.e., originating from the same patient or sexual partners) sequences and identified 18 different MgPa-13 region variants through comparative sequence analysis. Identical strains were detected at different anatomical sites of four female patients, which demonstrated colonization with the same strain. Further, identical strains were observed among partners (four dyads), corroborating previously observed transmission of M. genitalium between sexual partners (14, 17, 27).
We compared San Antonio area-originating MgPa-13 nucleotide sequences with all available clinical sequences collected from different locations. Fifty-six different MgPa-13 variants were recently recognized among 267 sequences found in positive specimens from nine countries (14). Eight different strains were identified among female commercial sex workers in Nairobi, Kenya (17). Alignment of DNA sequences revealed identical MgPa-13 variants in different groups of patients, implying the existence of several common sequence types worldwide. Alongside these common sequence variants, location-specific sequences were represented.
Although MgPa-13 regions demonstrated extensive DNA sequence variability, amino acid substitutions seemed to involve only a limited number of amino acids (Fig. 2 and 3). In addition, numerous silent mutations were observed (38% of observed codon mutations). For example, comparison of deduced amino acid sequences indicated that all detected leucine codon changes (L78, L90, and L115) were identical and silent. Furthermore, conservation of N99, I100, and K120 in their positions was observed (i.e., in each case, except for a single missense mutation, all substitutions were identical and silent) (Fig. 2 and 3). Additionally, it appears that several of the silent mutations were found together in the same sequence variant.
As mentioned above, missense mutations involved a limited number of amino acids (Fig. 2 and 3), and simultaneous occurrence of several missense mutations was also detected in strains of different geographic origin. As a result, several distinct sequence variants of adhesin MgPa were identified (Fig. 3).
In the majority of synonymous mutations, except for L115, codons (as found in G37) were replaced by less abundant codons (Table 1). Thus, even though the amino acid sequence of MgPa remains unchanged, codon mutations could affect expression of the MgPa protein. Effects of silent mutations on protein folding have been reported (6, 24). The function of MgPa as a major adhesin and its antigenicity have been demonstrated, but the exact positioning of the protein in the mycoplasma membrane, as well as the role of individual domains or regions, has not been detailed. The impact of detected mutations on structural or functional properties of MgPa necessitates additional investigation.
Interestingly, the mgpB gene of M. genitalium and the adhesin P1 coding gene MPN141 of M. pneumoniae share much in common. Both genes contain repetitive sequences with multiple copies spread throughout the chromosome, and both genes contain a unique region within the proximal ∼500 nucleotides (7, 31, 32). Still, no sequence variability within this region has been observed with M. pneumoniae (37). In contrast to the recent findings of extensive sequence variability in M. genitalium (17, 18, 27, 34), the existence of two highly conserved types of M. pneumoniae P1 gene sequences has been demonstrated repeatedly (9). The existence of these two types of the P1 gene (type 1 and type 2) was shown to be an excellent marker for numerous other variations within their genomes, some of which likely lead to virulence differences observed among strains (8). It is possible that well-documented shifts of clinical M. pneumoniae isolates in different locations over time could be a result of type-associated differences (23). One hypothesis speculates that type-specific antibodies render protection against the causative type strain, leading to replacement by the other strain type. It is unclear whether any correlation exists between the particularly expressed MgPa region and M. genitalium virulence. As increased numbers of sequence variants arise, possible connections between MgPa-13 sequence variants and clinical symptoms, host immunoresponsiveness, and/or persistence of infection need to be evaluated. Accumulation and analysis of additional MgPa-13 region sequence data should further enhance studies of M. genitalium infections, evaluation of treatment effectiveness, and understanding of its epidemiology, transmission, pathogenicity, and phylogeny.
Acknowledgments
The project described was supported by award number U19AI045429 from the National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
We thank Caleb Herrera for technical assistance in specimen analysis and amplification of target sequences. We express gratitude to T. R. Kannan for help with writing this article.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Anagrius, C., B. Lore, and J. S. Jensen. 2005. Mycoplasma genitalium: prevalence, clinical significance, and transmission. Sex. Transm. Infect. 81458-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baseman, J. B., M. Cagle, J. E. Korte, C. Herrera, W. G. Rasmussen, J. G. Baseman, R. Shain, and J. M. Piper. 2004. Diagnostic assessment of Mycoplasma genitalium in culture-positive women. J. Clin. Microbiol. 42203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradshaw, C. S., M. Y. Chen, and C. K. Fairley. 2008. Persistence of Mycoplasma genitalium following azithromycin therapy. PLoS ONE 3e3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clausen, H. F., J. Fedder, M. Drasbek, P. K. Nielsen, B. Toft, H. J. Ingerslev, S. Birkelund, and G. Christiansen. 2001. Serological investigation of Mycoplasma genitalium in infertile women. Hum. Reprod. 161866-1874. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, C. R., L. E. Manhart, E. A. Bukusi, S. Astete, R. C. Brunham, K. K. Holmes, S. K. Sinei, J. J. Bwayo, and P. A. Totten. 2002. Association between Mycoplasma genitalium and acute endometritis. Lancet 359765-766. [DOI] [PubMed] [Google Scholar]
- 6.Cortazzo, P., C. Cervenansky, M. Marin, C. Reiss, R. Ehrlich, and A. Deana. 2002. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem. Biophys. Res. Commun. 293537-541. [DOI] [PubMed] [Google Scholar]
- 7.Dallo, S. F., A. Chavoya, C. J. Su, and J. B. Baseman. 1989. DNA and protein sequence homologies between the adhesins of Mycoplasma genitalium and Mycoplasma pneumoniae. Infect. Immun. 571059-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumke, R., I. Catrein, R. Herrmann, and E. Jacobs. 2004. Preference, adaptation and survival of Mycoplasma pneumoniae subtypes in an animal model. Int. J. Med. Microbiol. 294149-155. [DOI] [PubMed] [Google Scholar]
- 9.Dumke, R., I. Catrein, E. Pirkil, R. Herrmann, and E. Jacobs. 2003. Subtyping of Mycoplasma pneumoniae isolates based on extended genome sequencing and on expression profiles. Int. J. Med. Microbiol. 292513-525. [DOI] [PubMed] [Google Scholar]
- 10.Falk, L., H. Fredlund, and J. S. Jensen. 2005. Signs and symptoms of urethritis and cervicitis among women with or without Mycoplasma genitalium or Chlamydia trachomatis infection. Sex. Transm. Infect. 8173-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270397-403. [DOI] [PubMed] [Google Scholar]
- 12.Haggerty, C. L. 2008. Evidence for a role of Mycoplasma genitalium in pelvic inflammatory disease. Curr. Opin. Infect. Dis. 2165-69. [DOI] [PubMed] [Google Scholar]
- 13.Hamasuna, R., Y. Osada, and J. S. Jensen. 2007. Isolation of Mycoplasma genitalium from first-void urine specimens by coculture with Vero cells. J. Clin. Microbiol. 45847-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hjorth, S. V., E. Bjornelius, P. Lidbrink, L. Falk, B. Dohn, L. Berthelsen, L. Ma, D. H. Martin, and J. S. Jensen. 2006. Sequence-based typing of Mycoplasma genitalium reveals sexual transmission. J. Clin. Microbiol. 442078-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, P. C., W. A. Clyde, Jr., and A. M. Collier. 1984. Conservation of pathogenic mycoplasma antigens. Isr. J. Med. Sci. 20:916-919. [PubMed] [Google Scholar]
- 16.Hu, P. C., U. Schaper, A. M. Collier, W. A. Clyde, Jr., M. Horikawa, Y. S. Huang, and M. F. Barile. 1987. A Mycoplasma genitalium protein resembling the Mycoplasma pneumoniae attachment protein. Infect. Immun. 551126-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iverson-Cabral, S. L., S. G. Astete, C. R. Cohen, E. P. Rocha, and P. A. Totten. 2006. Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences. Infect. Immun. 743715-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iverson-Cabral, S. L., S. G. Astete, C. R. Cohen, and P. A. Totten. 2007. mgpB and mgpC sequence diversity in Mycoplasma genitalium is generated by segmental reciprocal recombination with repetitive chromosomal sequences. Mol. Microbiol. 6655-73. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, J. S. 2004. Mycoplasma genitalium: the aetiological agent of urethritis and other sexually transmitted diseases. J. Eur. Acad. Dermatol. Venereol. 181-11. [DOI] [PubMed] [Google Scholar]
- 20.Jensen, J. S., E. Bjornelius, B. Dohn, and P. Lidbrink. 2004. Use of TaqMan 5′ nuclease real-time PCR for quantitative detection of Mycoplasma genitalium DNA in males with and without urethritis who were attendees at a sexually transmitted disease clinic. J. Clin. Microbiol. 42683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, J. S., H. T. Hansen, and K. Lind. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J. Clin. Microbiol. 34286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, J. S., S. A. Uldum, J. Sondergard-Andersen, J. Vuust, and K. Lind. 1991. Polymerase chain reaction for detection of Mycoplasma genitalium in clinical samples. J. Clin. Microbiol. 2946-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenri, T., N. Okazaki, T. Yamazaki, M. Narita, K. Izumikawa, M. Matsuoka, S. Suzuki, A. Horino, and T. Sasaki. 2008. Genotyping analysis of Mycoplasma pneumoniae clinical strains in Japan between 1995 and 2005: type shift phenomenon of M. pneumoniae clinical strains. J. Med. Microbiol. 57469-475. [DOI] [PubMed] [Google Scholar]
- 24.Komar, A. A., T. Lesnik, and C. Reiss. 1999. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 462387-391. [DOI] [PubMed] [Google Scholar]
- 25.Korte, J. E., J. B. Baseman, M. P. Cagle, C. Herrera, J. M. Piper, A. E. Holden, S. T. Perdue, J. D. Champion, and R. N. Shain. 2006. Cervicitis and genitourinary symptoms in women culture positive for Mycoplasma genitalium. Am. J. Reprod. Immunol. 55265-275. [DOI] [PubMed] [Google Scholar]
- 26.Lind, K., B. O. Lindhardt, H. J. Schutten, J. Blom, and C. Christiansen. 1984. Serological cross-reactions between Mycoplasma genitalium and Mycoplasma pneumoniae. J. Clin. Microbiol. 201036-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, L., J. S. Jensen, L. Myers, J. Burnett, M. Welch, Q. Jia, and D. H. Martin. 2007. Mycoplasma genitalium: an efficient strategy to generate genetic variation from a minimal genome. Mol. Microbiol. 66220-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, L., and D. H. Martin. 2004. Single-nucleotide polymorphisms in the rRNA operon and variable numbers of tandem repeats in the lipoprotein gene among Mycoplasma genitalium strains from clinical specimens. J. Clin. Microbiol. 424876-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manhart, L. E., C. W. Critchlow, K. K. Holmes, S. M. Dutro, D. A. Eschenbach, C. E. Stevens, and P. A. Totten. 2003. Mucopurulent cervicitis and Mycoplasma genitalium. J. Infect. Dis. 187650-657. [DOI] [PubMed] [Google Scholar]
- 30.Mena, L., X. Wang, T. F. Mroczkowski, and D. H. Martin. 2002. Mycoplasma genitalium infections in asymptomatic men and men with urethritis attending a sexually transmitted diseases clinic in New Orleans. Clin. Infect. Dis. 351167-1173. [DOI] [PubMed] [Google Scholar]
- 31.Morrison-Plummer, J., D. H. Jones, K. Daly, J. G. Tully, D. Taylor-Robinson, and J. B. Baseman. 1987. Molecular characterization of Mycoplasma genitalium species-specific and cross-reactive determinants: identification of an immunodominant protein of M. genitalium. Isr. J. Med. Sci. 23453-457. [PubMed] [Google Scholar]
- 32.Morrison-Plummer, J., A. Lazzell, and J. B. Baseman. 1987. Shared epitopes between Mycoplasma pneumoniae major adhesin protein P1 and a 140-kilodalton protein of Mycoplasma genitalium. Infect. Immun. 5549-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musatovova, O., S. Dhandayuthapani, and J. B. Baseman. 2003. Transcriptional starts for cytadherence-related operons of Mycoplasma genitalium. FEMS Microbiol. Lett. 22973-81. [DOI] [PubMed] [Google Scholar]
- 34.Musatovova, O., C. Herrera, and J. B. Baseman. 2006. Proximal region of the gene encoding cytadherence-related protein permits molecular typing of Mycoplasma genitalium clinical strains by PCR-restriction fragment length polymorphism. J. Clin. Microbiol. 44598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, J. D., and J. S. Jensen. 2006. Mycoplasma genitalium as a sexually transmitted infection: implications for screening, testing, and treatment. Sex. Transm. Infect. 82269-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shain, R. N., J. M. Piper, E. R. Newton, S. T. Perdue, R. Ramos, J. D. Champion, and F. A. Guerra. 1999. A randomized, controlled trial of a behavioral intervention to prevent sexually transmitted disease among minority women. N. Engl. J. Med. 34093-100. [DOI] [PubMed] [Google Scholar]
- 37.Su, C. J., S. F. Dallo, and J. B. Baseman. 1990. Molecular distinctions among clinical isolates of Mycoplasma pneumoniae. J. Clin. Microbiol. 281538-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tosh, A. K., B. Van Der Pol, J. D. Fortenberry, J. A. Williams, B. P. Katz, B. E. Batteiger, and D. P. Orr. 2007. Mycoplasma genitalium among adolescent women and their partners. J. Adolesc. Health 40412-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Totten, P. A., M. A. Schwartz, K. E. Sjostrom, G. E. Kenny, H. H. Handsfield, J. B. Weiss, and W. L. Whittington. 2001. Association of Mycoplasma genitalium with nongonococcal urethritis in heterosexual men. J. Infect. Dis. 183269-276. [DOI] [PubMed] [Google Scholar]