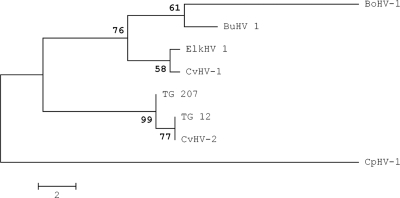
FIG. 2.
Phylogenetic relationships between the UL27 gene sequences of two samples obtained from trigeminal ganglia, TG_12 and TG_207, and those of other ruminant alphaherpesviruses. TG_207 was identical to eight of the sequences obtained in this study, while TG_12 was identical to the remaining seven sequences. The sequences retrieved from GenBank (with accession numbers in parentheses) were BoHV-1 (NC001847.1), CvHV-1 (AF078729.2), CvHV-2 (AF078727.2), BuHV-1 (EF624476.1), ElkHV-1 (EF624478.1), and CpHV-1 (AF078728.2). The maximum-parsimony method was used. Tree branches corresponding to partitions reproduced in <50% of bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to each branch. The maximum-parsimony tree was obtained by using the close-neighbor-interchange algorithm with a search level of 3, in which the initial trees were obtained by the random addition of sequences (10 replicates). The tree is drawn to scale. Branch lengths were calculated using the average pathway method; the unit is the number of changes over the whole sequence. There were 262 positions in the final data set, 18 of which were informative of parsimony.