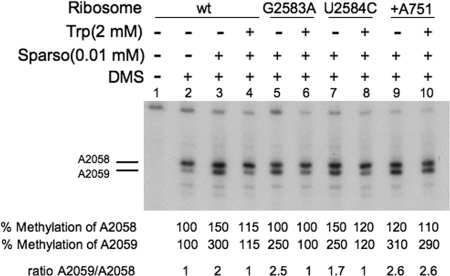
FIG. 3.
Action of the antibiotic sparsomycin on different isolated TnaC-tRNA-ribosome-mRNA complexes. Complexes isolated as indicated in the legend of Fig. 3 were prepared using the biotinylated tnaC mRNA designated for each panel. The isolated complexes were preincubated for 5 min with (+) or without (−) sparsomycin (Sparso) at 37°C. They were then incubated for five additional minutes at the same temperature in the presence (+) or absence (−) of Trp. Finally, the mixtures were exposed to the methylating reagent DMS to modify the A2058 and A2059 adenine nucleotides of 23S rRNA (positions indicated by horizontal lines). A sample was incubated in the absence of DMS as a methylation control. The rRNA isolated from the final mixtures was used to perform primer extension analyses. The percent methylation shown corresponds to the ratio of the cpm detected for each A2058 and A2059 band divided by the cpm for the corresponding band obtained with the control sample of ribosome-tnaC mRNA without tryptophan. wt, wild type.