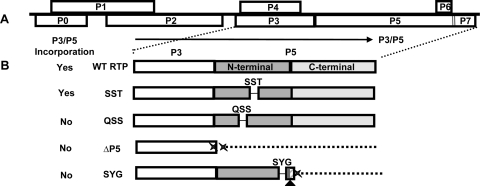
FIG. 1.
Genome organization of PLRV and illustration of P5 mutant viruses. (A) The 6-kb monopartite, positive-sense, single-stranded RNA genome consisting of eight ORFs. ORFs P3 to P5, which are involved in structure and movement, are conserved among luteoviruses and are expressed from subgenomic RNA-1. Virions are composed mainly of P3 coat proteins and a small number of P3/P5 readthrough proteins, which are anchored into the virion by the P3 moiety. The P5 domain is exposed on the surface and has a conserved N-terminal region and a variable C-terminal region. (B) Illustration of the P3/P5 translation of the mutant viruses described in this study. Deletion of amino acids is specified as a line, and a lack of protein expression is shown as a dotted line. WT PLRV with expression of the entire P3/P5 is depicted. For the SST and QSS mutant viruses, the 9 nt comprising the SST sequence (nt 4531 to 4539) and the 9-nt QSS sequence (nt 4438 to 4446) were deleted, respectively, while full-length P3/P5 translation was maintained. The resulting P3/P5 protein was incorporated into the virion for the SST mutant virus but not for the QSS mutant virus. The ΔP5 mutant virus contains two stop codons at the end of the P3 sequence followed by a 100-nt deletion in P5 (double Xs); no P3/P5 translation occurs. The SYG mutation is located 18 amino acids from the C terminal region. In addition to the 9-nt deletion to create the SYG deletion (nt 4828 to 4836), a single nucleotide was deleted 14 nt downstream (nt 4850) (triangle). This created a frameshift, which altered the subsequent amino acids (diagonal lines) and eventually produced a stop codon (X) 17 nt downstream from the single-nucleotide deletion. As a result, a truncated P3/P5 was translated. ΔP5 and SYG mutant viruses do not have P3/P5 incorporated into their virions.