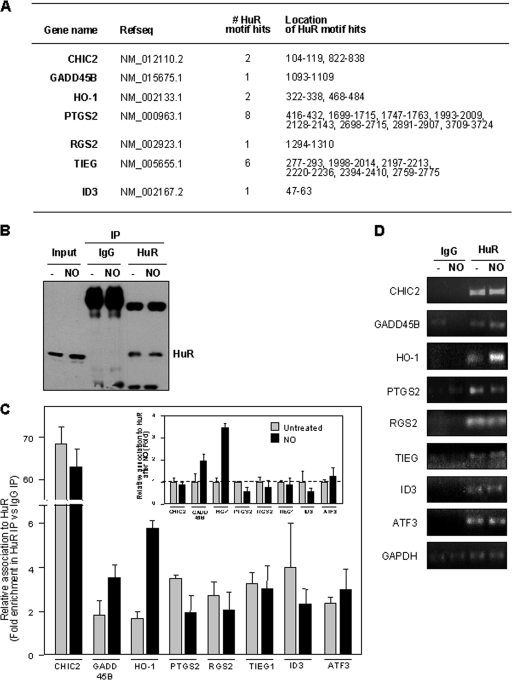
FIG. 4.
(A) Predicted hits for a signature HuR motif. The numbers and locations within of HuR motif hits within the 3′UTR for each transcripts are indicated. (B) Western blot analysis of HuR levels in lysates from untreated and NO-treated cells before IP (Input) (10 μg per lane) and after IP (from 100-μg aliquots) with IgG and with an anti-HuR antibody (25% of IP material loaded). (C) Relative association of HuR with the transcripts shown in IMR-90 cells that were either left untreated or treated with NO (0.5 mM for 2.5 h). Following HuR IP (and parallel control IgG IP), RNA isolation, RT-qPCR analysis, and normalization to GAPDH mRNA levels in each IP sample, the abundance of each mRNA in HuR IP compared with that in IgG IP was calculated. Data (means ± SD from three independent experiments) are plotted as relative enrichment in HuR IP versus IgG IP (main graph) and also as the levels of enrichment in NO relative to untreated populations (inset graph). (D) The enrichment of each mRNA in each IP sample was also visualized by RT followed by PCR amplification for 22 cycles for all mRNAs except for GAPDH, which was amplified for 28 cycles; PCR products were visualized in ethidium bromide-stained agarose gels.