Abstract
Recently, autophagy has emerged as a critical process in the control of T-cell homeostasis. Given the pivotal role of NF-κB in the signaling events of T cells, we have analyzed and unveiled a conserved NF-κB binding site in the promoter of the murine and human BECN1 autophagic gene (Atg6). Accordingly, we demonstrate that the NF-κB family member p65/RelA upregulates BECN1 mRNA and protein levels in different cellular systems. Moreover, p65-mediated upregulation of BECN1 is coupled to increased autophagy. The newly identified κB site in the BECN1 promoter specifically interacts with p65 both in vitro and in living Jurkat cells upon phorbol myristate acetate (PMA)-ionomycin stimulation, where p65 induction is coupled to BECN1 upregulation and autophagy induction. Finally, anti-CD3- and PMA-ionomycin-mediated activation of T-cell receptor signaling in peripheral T cells from lymph nodes of healthy mice results in an upregulation of BECN1 expression that can be blocked by the NF-κB inhibitor BAY 11-7082. Altogether, these data suggest that autophagy could represent a novel route modulated by p65 to regulate cell survival and control T-cell homeostasis.
Normal cellular development and growth depend on a finely tuned balance between protein synthesis and degradation. Eukaryotic cells exploit two major routes for protein degradation: autophagy and the ubiquitin-proteasome system. Autophagy is a highly conserved catabolic process whereby long-lived proteins and organelles are engulfed in double-membrane structures called autophagosomes and targeted to the lysosome for degradation and ATP production (33). Autophagy was first characterized in yeast as a process used by cells to survive metabolic stress (59). Many of the autophagic executor genes (ATGs) have been discovered in yeast, and recently a number of mammalian orthologues have been identified. At the molecular level, the autophagic pathway is well conserved and requires several proteins acting in concert at different stages for proper autophagosome formation. During the first steps of autophagosome formation, BECN1/Atg6 acts in association with PI3KIII/Vps34 as a platform that recruits activators and inhibitors of autophagy in order to finely regulate autophagosome formation (43). Two ubiquitin-like systems are fundamental for autophagosome enlargement and maturation downstream of BECN1. A first ubiquitin-like system is required for the formation of the Atg5-Atg12 complex, which contributes to autophagosomal membrane elongation (30, 39). The second ubiquitin-like system conjugates LC3 protein to phosphoethanolamine, allowing its incorporation into the autophagosomal membrane and subsequent autophagosome formation (56).
In mammalian cells, metabolic stress and genotoxic stimuli, including ceramide and tamoxifen treatment, have been shown to trigger both apoptotic cell death and autophagy (51). A number of emerging pieces of evidence point to a prosurvival role for autophagy. Inactivation of the essential autophagy gene BECN1 results in apoptotic cell death in different models (8, 11, 55). Autophagy has been shown to represent a temporary survival pathway that counteracts metabolic stress in tumors with defective apoptosis, and it is required for the maintenance of energy homeostasis in healthy cells during postnatal starvation (29). Intriguingly, impairment of apoptosis often leads to autophagy-dependent survival of cells subjected to metabolic stress, suggesting that tumor cells, at least in some instances, could avoid cell death through autophagy (12, 37). Other pieces of evidence, on the other hand, indicate that autophagy could represent a tumor suppressor pathway; indeed, heterozygous disruption of the BECN1 gene in mice results in enhanced tumor formation (48). Accordingly, BECN1 is monoallelically deleted in many human breast, prostate, and ovarian cancers (1). Moreover, when allowed to reach completion, autophagy can ultimately result in cell death (type II programmed cell death) (34), suggesting that autophagy activation may lead to different outcomes, depending on the cell genetic background and the stimulus used.
Concerning the transcriptional regulation of autophagy, a complex picture has recently emerged. Several reports indicate that different transcription factors can regulate the expression of ATGs in a cell context- and stimulus-specific manner. FoxO3 has been shown to directly regulate the expression of Gabarapl1, LC3b, and Atg12l in myotubes (66). Other findings have confirmed the ability of HIF-1α to either indirectly regulate both BECN1/Atg6 and Atg5 (4, 64) or directly bind the BNIP3 promoter (58). Interestingly, a most recent work showed that E2F1 binds the Atg1/ULK1, MAPLC3, and DRAM1 promoters, increasing their expression and regulating autophagy (44). Besides E2F1, p53 is induced in response to genotoxic stress and recently it has been shown to have a dual role in the regulation of autophagy. In fact, while nuclear p53 directly induces DRAM1 and autophagy (9), cytoplasmic p53 negatively regulates autophagosome formation (32, 57). Interestingly, several studies have highlighted a cross talk between p65 and either p53 (49) or HIF-1α. Intriguingly, HIF-1α has been recently proven to be a direct target of p65 (60) and it has already been established that these transcription factors share several target genes (36). The relationship between NF-κB signaling and autophagy regulation seems puzzling. Indeed, NF-κB has emerged as a negative regulator of autophagy as induced by tumor necrosis factor alpha (TNF-α), reactive oxygen species, and starvation in some cell lines (16, 18). On the other hand, a number of indirect pieces of evidence suggest an activating role for NF-κB in autophagy. NF-κB is positively regulated by the TSC2/mTor inhibitor (20), and the NF-κB inhibitor IκBα can be degraded through a lysosomal route following starvation in CHO cells (10). Remarkably, a number of NF-κB-activating stimuli, including ceramide, TNF-α, tamoxifen, and endoplasmic reticulum (ER) stress, can also induce autophagy (26, 27, 50, 51, 54).
The NF-κB prototype, the p65/p50 dimer, is responsible for both B-cell and T-cell survival and activation, as shown by studies with p50-deficient mice and RelA−/− T cells, respectively (17, 53). Interestingly, autophagy was also shown to play an essential role in T-lymphocyte survival and function (45) but also to be instrumental in growth factor withdrawal cell death (35).
Activation of NF-κB often results in contradictory outcomes, as exemplified in T cells, where NF-κB can either promote cell survival by transactivating antiapoptotic genes such as Bcl-2 and inhibitors of apoptosis or trigger cell death by inducing FasL expression during activation-induced cell death (67). NF-κB-mediated transactivation of antiapoptotic target genes has confirmed a prosurvival and pro-oncogenic role for this transcription factor. Nevertheless, keratinocytes from RelA−/− mice have been shown to hyperproliferate in vitro (65), suggesting that the final biological outcome of NF-κB activation is strictly cell context and stimulus dependent.
In this work, we show that BECN1 is a novel p65 target gene and present evidence that blockage of p65 signaling leads to a decrease in BECN1 transcription. As a consequence, autophagy is impaired following p65 depletion. Our findings establish a link between p65 and autophagy, suggesting that p65 is required for BECN1 expression. Finally, we provide evidence suggesting that p65-mediated regulation of BECN1 could be important for T-cell activation and proliferation, highlighting a novel mechanism whereby p65 could promote cell survival.
MATERIALS AND METHODS
Reagents and plasmids.
C2 ceramide, tamoxifen, phorbol myristate acetate (PMA), BAY 11-75082, G418, and leupeptin were purchased from Sigma-Aldrich. Ponasterone A, Zeocin, and Oligofectamine were from Invitrogen. FuGENE 6 was from Roche Diagnostics. Control small interfering RNA (siRNA) GUGACCAGCGAAUACCUGU (siCONT/SiCtrl) (62), BECN1 siRNA (GAUUGAAGACACAGGAGGCTT) (6), and p65/RelA siRNA (GCCCUAUCCCUUUACGUCATT) (40) were purchased from MWG.
pIND and pVgRXR were from Invitrogen. The pGL3 vector was from Promega. pIND p65/RelA, pIND IKBαSR, pGL3CHET4, and pGL3SCHET4 were generated by subcloning the relevant PCR products into the vectors of interest.
Cell lines and treatments.
The U2OS, HEK293, and HeLa cell lines were routinely cultured at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 mg/ml). Jurkat E6.1 cells were grown in suspension in RPMI medium supplemented with 10% (vol/vol) fetal bovine serum, penicillin (100 U/ml), streptomycin (100 mg/ml), and glutamine (200 mM). U2OS derivative cell lines were obtained by cotransfection of pVgRXR and pIND p65 or pIND IκBaSR at a ratio of 1:9. Selection was performed with Zeocin (Invitrogen) and G418 (Sigma).
Immunological procedures and quantifications.
Western blot analysis and immunofluorescence assays were performed according to standard procedures with anti-BECN1 sc-11427, NF-κB p65 antibody sc-372X, and anti-p50 sc-1190 from Santa Cruz Biotechnology and antiactin polyclonal antibody and antihemagglutinin monoclonal antibody from Sigma. Anti-LC3 antibody was produced by immunization of a rabbit with glutathione S-transferase-tagged recombinant LC3 (rLC3) protein. Antisera were affinity purified with six-His-tagged rLC3 protein covalently coupled to Affi-Prep 10 matrix (Bio-Rad). ImageJ software (NIH) was used to quantify gel bands. Fluorescence-activated cell sorter (FACS) analysis and reagents for monitoring T-cell activation were previously described (46).
Statistical analysis.
Results are expressed as means ± standard deviations of at least three independent experiments. Statistical analysis (unless otherwise indicated) was performed with a t test with the level of significance set at P < 0.05.
Semiquantitative reverse transcription (RT)-PCR of BECN1 and quantitative RT-PCR (qRT-PCR).
RNA from 2 × 106 HEK293 cells per experiment was used. Semiquantitative RT-PCR was performed as previously described (51) with the same set of BECN1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers.
qRT-PCR was performed as follows. RNA extraction and retrotranscription were performed with the same procedure used for the semiquantitative RT-PCR experiments. Fifty nanograms of each sample was used for mRNA quantitation. BECN1 gene expression was detected by qRT-PCR assay. Quantification of BECN1 mRNA was achieved by means of the STEP One Real-Time PCR System (Applied Biosystems [AB]) with TaqMan Universal Master Mix (AB). qRT-PCR was based upon the TaqMan fluorogenic detection system (AB). The primer and probe sequences used for qRT-PCR with BECN1 were as follows: primers, 5′-GTCCTCCCCGTATCATACCATTC-3′ and 5′-ACGATGGTAAAGGGAGGGAAGT-3′; TaqMan 6-carboxyfluorescein probe, 5′-CAGTGGCGGCTCC-3′. A predeveloped human GAPDH probe was purchased from AB. Triplicate RT-PCRs were prepared for each sample. The ΔCT method was applied by using the GAPDH cycle threshold for normalization to evaluate relative BECN1 mRNA expression. A one-tailed Student t test was used to assess the statistical significance of the observed mRNA differences in qPCR experiments. Differences were considered significant at P < 0.05.
In silico analysis.
In silico analysis was performed with CONFAC (28). Concerning the data set of autophagic genes, the upstream sequence length was fixed at 500 bp, matrix similarity was fixed at 0.85, and core similarity was fixed at 0.95. The Vertebrate matrix was used, and Repeatmasker was applied.
The CHET4 sequence was analyzed with TRANSFAC Professional (38).
Electrophoretic mobility shift assays (EMSAs) and chromatin immunoprecipitation (ChIP).
EMSAs were performed as described previously (14). For ChIP analysis, 108 Jurkat cells per experimental point were used. ChIP was performed according to the protocol of A. Kouskouti and I. Kyrmizi, which is available online (http://www.epigenome-noe.net/researchtools/pdfs/p10.pdf). ChIP was performed with anti-p65 antibody sc-372X (Santa Cruz); preimmune serum was used as an “unrelated antibody.” Chromatin from cross-linked cells was sheared by sonication (three times, 30 s on/30 s off, medium power; Diagenode Bioruptor). Precipitated DNAs were analyzed by PCR (35 cycles). Primers for the human IκBα promoter and the β-actin promoter were previously reported (63). Specific primers for the A κB site in the BECN1 promoter are 5′-CCCGTATCATACCATTCCTAG-3′ and 5′-GAAACTCGTGTCCAGTTTCAG-3′. qRT-PCR with the STEP One Real-Time PCR System was performed in triplicate with Sybr green PCR Master Mix (AB) to assess the interaction of p65 with the A κB site in the BECN1 promoter, the IκBα promoter, and the β-actin promoter.
RESULTS
In silico analysis of the BECN1 promoter.
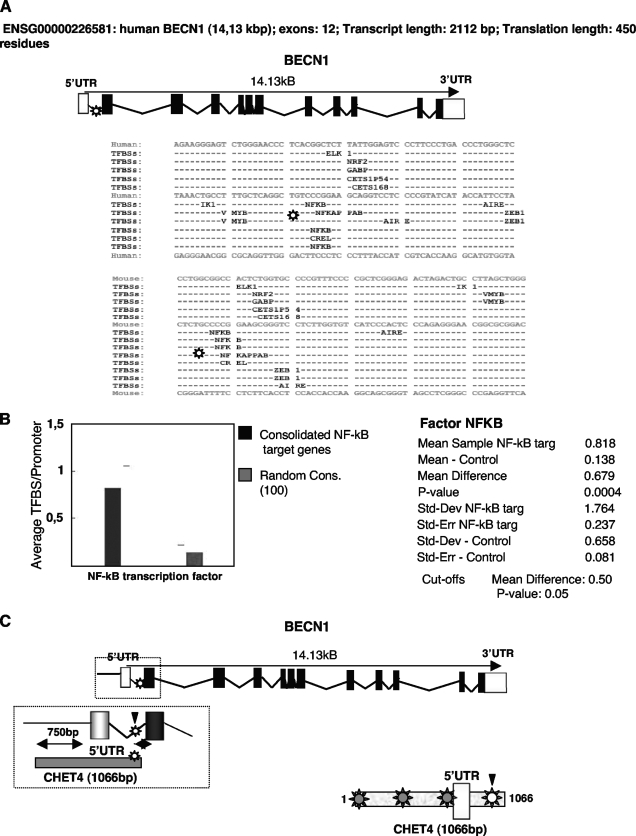
To explore the transcriptional regulation of the autophagic process, we performed an in silico analysis with the CONFAC tool (28) to investigate whether any transcription factor binding site was conserved in the promoters of a list of genes classified as “autophagic” by gene ontology in humans and mice (see Fig. S1 in the supplemental material). CONFAC analysis revealed that no common transcription factor binding site was conserved in the promoter regions including 500 bp upstream of the transcription start site and up to 15 kbp of the first intron of the submitted genes. However, considering the outputs relative to each gene, three putative overlapping NF-κB binding sites (κB sites) were found inside the first intron of the BECN1 promoter in both species (Fig. 1A) (25). Subsequently, in order to verify the results obtained for the BECN1 promoter, a list of NF-κB target genes (see Fig. S2 in the supplemental material), including BECN1, was submitted to CONFAC. A Mann-Whitney test performed with the set of NF-κB target genes and a set of 100 randomly selected genes from RefSeq (see Fig. S3 in the supplemental material) demonstrated that κB sites in the promoters of NF-κB target genes were statistically significantly overrepresented. In particular, the analyzed data set showed an average number of κB site occurrences per promoter of 0.8, in contrast to the 0.1 of the negative control gene list, confirming that the presence of three κB sites in the BECN1 promoter was statistically significant (Fig. 1B) and suggesting that BECN1 could represent a novel NF-κB target gene. To further explore this hypothesis, a DNA segment containing 1.1 kbp of the human BECN1 promoter (CHET4), previously cloned as an E2F binding region (61), was analyzed with the TRANSFAC Professional tool (see Fig. S4 in the supplemental material) (38), confirming the presence of the κB site predicted by CONFAC inside the first intron of the human BECN1 gene. Moreover, the same analysis highlighted the existence of three other putative κB sites (Fig. 1C) in a more upstream region, further suggesting that NF-κB could regulate BECN1 expression.
FIG. 1.
In silico analysis of the BECN1 promoter. (A, top) genomic organization of the human (ENSG00000126581) and mouse (ENSMUSG00000035086) BECN1 genes according to Ensembl. (A, bottom) Exact position (as shown by asterisks) of the κB site predicted in the human and mouse BECN1 promoters by CONFAC analysis. (B) Mann-Whitney test results. CONFAC consolidated conserved transcription factor binding sites for the list of NF-κB target (targ) genes, including BECN1, and for a control list of 100 randomly selected control genes were compared. Statistical analysis parameters are shown on the right. (C) Exact positions (as shown by asterisks) of the κB sites located in a DNA segment containing 1.1 kbp (CHET4) of the human BECN1 promoter, according to TRANSFAC Professional. The κB site confirmed both by CONFAC and TRANSFAC is in white. (Top) Genomic organization of BECN1 with respect to the CHET4 sequence (boxed). UTR, untranslated region; Std, standard; Dev, deviation; Err, error; Cons., consolidated.
NF-κB/p65 induces the human BECN1 promoter.
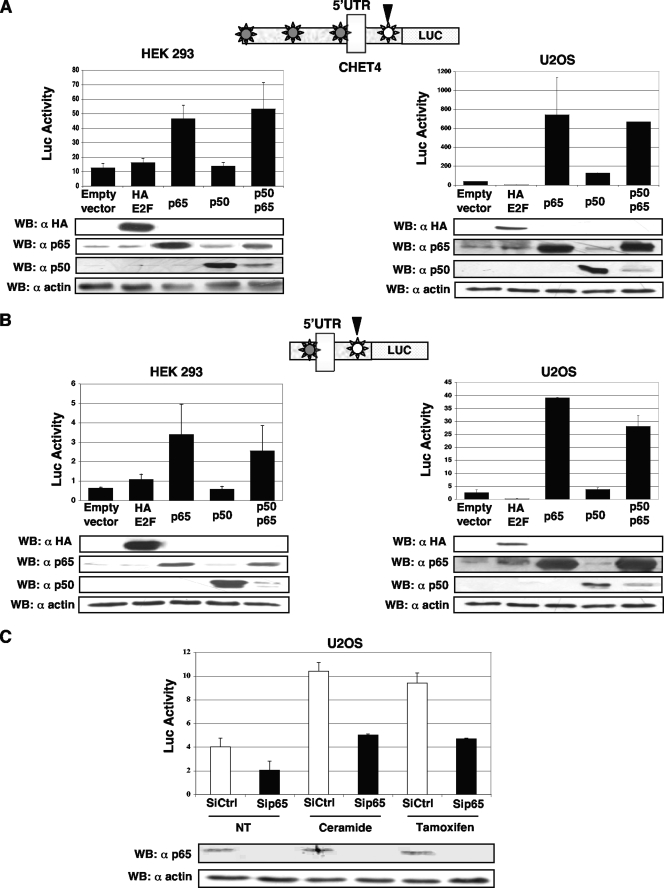
To investigate whether NF-κB indeed regulates BECN1 expression in living cells, the 1.1-kbp (CHET4) promoter region of the human BECN1 gene and a shorter segment (381 bp, SCHET4) encompassing the κB site predicted by CONFAC were inserted into the pGL3-basic luciferase reporter vector and the luciferase activities of the ectopic promoters were evaluated in response to the overexpression of p65 in the U2OS and HEK293 cell lines.
As shown in Fig. 2A, luciferase activity was significantly enhanced by p65 overexpression alone or in combination with p50 in both cell lines, while neither p50 nor E2F overexpression had any effect on human BECN1 promoter-driven luciferase transactivation. Interestingly, the effects of p65 on the shorter region of the human BECN1 promoter encompassing the κB site predicted by CONFAC were similar to those observed with the 1.1-kbp BECN1 promoter region (Fig. 2B), suggesting that the κB site found by CONFAC analysis is the main one responsible for the observed effects on the 1.1-kbp fragment of the BECN1 promoter.
FIG. 2.
p65 regulation of the human BECN1 promoter. (A) At the top is a schematic representation of the CHET4 region of the human BECN1 promoter. Asterisks represent the κB sites predicted by TRANSFAC. The κB site predicted by CONFAC is in white. The pGL3 firefly luciferase (Luc) reporter containing 1.1 kbp of the human BECN1 promoter was cotransfected into HEK293T and U2OS cells together with a Renilla luciferase plasmid and E2F-, p65-, p50-, p65-, and p50-expressing plasmids or a control empty vector, respectively. Dual-luciferase assays were performed 24 h after transfection. Data represent the means of at least four independent experiments, and error bars represent standard deviations. A Student t test indicated a significant difference from the control (P < 0.05). (Bottom) The levels of each overexpressed protein was monitored by Western blot (WB) analyses. (B) At the top is a representation of the SCHET4 region of the human BECN1 promoter. Asterisks represent the κB sites predicted by TRANSFAC. The κB site predicted by both CONFAC and TRANSFAC is in white. For luciferase assays, HEK293T and U2OS cells were cotransfected with pGL3SCHET4/Renilla luciferase and E2F-, p65-, p50-, p65-, and p50-expressing plasmids or a control empty vector. Data represent the means of at least four independent experiments, and error bars represent standard deviations. A Student t test indicated a significant difference from the control (P < 0.05). (Bottom) The level of each overexpressed protein was monitored by Western blot analyses. (C) p65 expression was selectively knocked down by siRNA in U2OS cells. At 48 h later, cotransfection of pGL3SCHET4 and a Renilla luciferase control (Ctrl) plasmid was performed. At 16 h after transfection, the cells were treated with 25 μM ceramide or 10 μM tamoxifen or not treated (NT) and 5 h later, cell lysates were prepared for luciferase activity quantification. Data represent the means of at least three independent experiments, and error bars represent standard deviations. A Student t test indicated a significant difference from the control (P < 0.05). (Bottom) Immunoblotting was carried out with the same lysates to monitor the levels of endogenous p65. UTR, untranslated region.
In order to address the role of endogenous p65 on human BECN1 promoter regulation, we assessed the effects of p65 (40) depletion on the transcriptional activity of the region inside the BECN1 promoter encompassing the putative κB site predicted by CONFAC in U2OS cells. As shown in Fig. 2C, depletion of endogenous p65 significantly decreased the promoter activity in U2OS cells, suggesting that p65 could play a positive role in the regulation of the BECN1 gene.
Previous studies have shown that ceramide and tamoxifen, two well-established autophagy inducers, may enhance BECN1 expression (51). Thus, we monitored the effect of endogenous p65 knockdown after 5 h of treatment with these drugs. In agreement with the previous published data, U2OS treatment with either ceramide or tamoxifen resulted in increased activity of the BECN1 promoter containing the consensus κB site (Fig. 2C). Notably, depletion of p65 strongly impairs the ability of both drugs to increase BECN1 promoter transactivation. Accordingly, previous reports demonstrated that ceramide can trigger p65 activation (5, 14) and that tamoxifen induction of both autophagy and BECN1 expression is mediated by ceramide (51). Altogether, these data indicate that induction of BECN1 gene expression by both ceramide and tamoxifen is probably mediated directly by the interaction of p65 with the κB site present in the first intron of BECN1.
p65 regulates BECN1 transcription.
Collectively, the data obtained by means of luciferase assays confirmed the in silico analysis and argued for involvement of p65 in the regulation of BECN1 transcription. To further address this hypothesis, we analyzed whether p65 can modulate BECN1 mRNA levels.
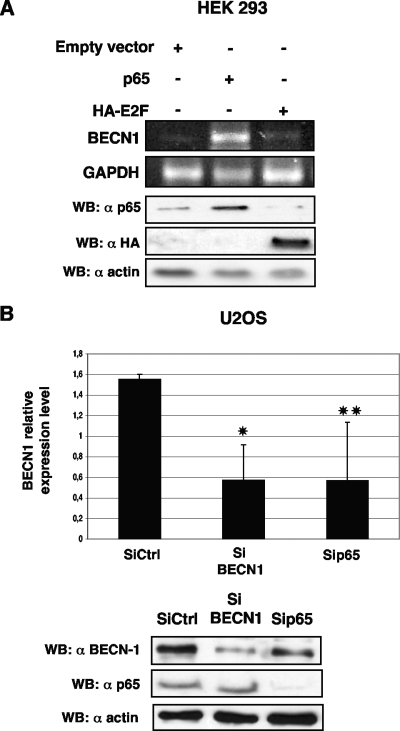
Semiquantitative RT-PCR assays were performed to detect the expression of BECN1 mRNA (51). As shown in Fig. 3A, p65 overexpression in HEK293 cells resulted in a significant increase in BECN1 transcription. In order to assess whether endogenous p65 could regulate basal BECN1 transcription, expression of p65 was knocked down with p65 siRNA in U2OS cells and the levels of BECN1 mRNA were quantified by qRT-PCR. As shown in Fig. 3B, depletion of p65 resulted in a significant decrease in BECN1 transcription. A scrambled siRNA (62) and a specific BECN1 siRNA (6) were used as negative and positive controls, respectively.
FIG. 3.
p65 regulation of BECN1 transcription. (A) Semiquantitative RT-PCR analysis of BECN1 and GAPDH mRNA expression in HEK 293T cells. Cells were transfected as indicated. RNAs were reverse transcribed and amplified with primers specific for BECN1 and GAPDH as described in Materials and Methods. The relative BECN1 mRNA level was normalized to the GAPDH mRNA level. The semiquantitative RT-PCR analysis was repeated three times, and the results of a representative experiment are shown. Proper expression of either p65 or E2F was assessed by immunoblotting. (B) qPCR analysis of BECN1 mRNA in U2OS cells. Cells were transfected with siRNAs as indicated. Equal amounts of total RNAs were reverse transcribed and amplified with a specific TaqMan BECN1 6-carboxyfluorescein probe and with a TaqMan GAPDH VIC probe as described in Materials and Methods. A Student t test was used to assess the statistical significance of the observed BECN1 mRNA differences in qPCR experiments. *, P < 0.005; **, P < 0.05. Immunoblot analysis confirmed the proper knockdown of either BECN1 or p65. WB, Western blotting; Ctrl, control.
Hence, we can argue that p65 positively regulates BECN1 transcription, probably due to its ability to directly act on the BECN1 promoter.
p65 regulates BECN1 protein expression levels in different cellular systems.
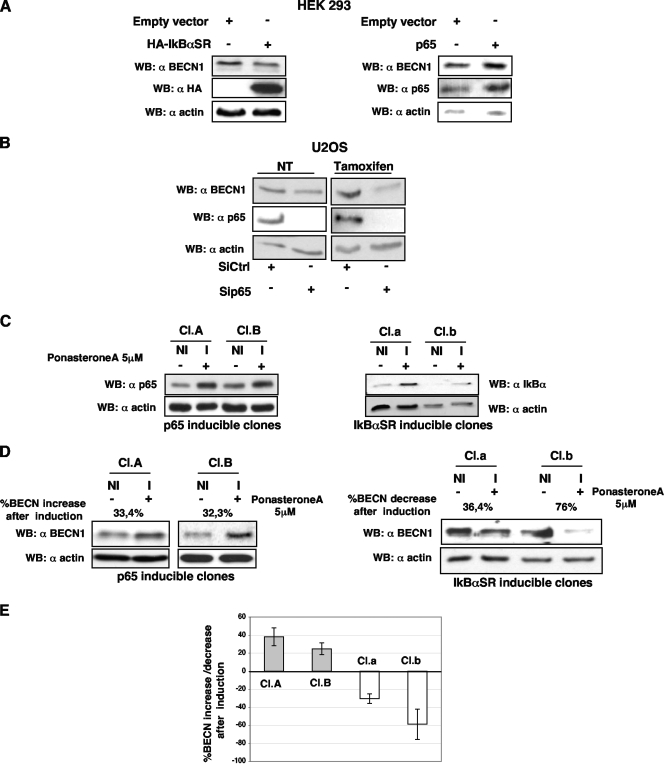
Since we demonstrated that p65 positively regulates BECN1 transcription, we expected a consequent increase in BECN1 protein levels. Therefore, the effects of either p65 or IκBαSR overexpression were assessed in HEK293 cells. IκBαSR protein behaves like a superrepressor of the classical NF-κB pathway since two serine residues (S32A/S36A) have been mutated, so that it cannot be targeted to proteasomal degradation. Consistent with the data obtained from the luciferase and semiquantitative RT-PCR assays, following p65 or IκBαSR overexpression, the expression levels of BECN1 protein in HEK293 cells were slightly but reproducibly increased or decreased, respectively (Fig. 4A). Furthermore, knockdown of p65 expression in U2OS cells again led to BECN1 protein downregulation (Fig. 4B) both under basal conditions and more prominently following tamoxifen treatment.
FIG. 4.
BECN1 protein level regulation by p65. (A) Lysates from HEK293 cells overexpressing hemagglutinin-tagged human IκBαSR, p65, or an empty vector were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently analyzed by immunoblotting for BECN1 protein expression. (B) BECN1 protein levels were evaluated by immunoblotting in untreated and tamoxifen (10 μM, 5 h)-treated U2OS cells following p65 depletion. Ctrl, control. (C) Generation of U2OS cell lines inducible for either p65 or IκBSR. Inducible cell lines were obtained as described in Materials and Methods. Two clones for each kind of inducible U2OS cells were tested. Treatment with 5 μM ponasterone A (I) for 16 h significantly increased the expression of both p65 and IκBαSR in the selected clones with respect to that in uninduced (NI) cells. (D, left) U2OS clones A and B, inducible for p65, were treated with 5 μM ponasterone A (I) or left untreated (NI) for 6 h. Cell lysates were subjected to Western blot (WB) analysis for monitoring of BECN1 levels. (D, right) Induction of IκBSR clones a and b was preformed, and BECN1 levels were assessed by immunoblotting. (E) Quantification of the bands was performed with the ImageJ tool. The graph is representative of three independent experiments performed with each of the four clones. Percentages of BECN1 protein increase or decrease following ponasterone A treatment are reported.
In order to overcome the engagement of an IκBα negative feedback that could reasonably mask the effect of p65 on the BECN1 protein level, two U2OS-derivative cell lines inducible for the expression of either p65 or IκBαSR were generated. Two representative clones for each inducible construct were used for further analysis (Fig. 4C). As shown in Fig. 4D (left side) and E, induction of p65 expression led to a significant enhancement of BECN1 protein levels in these experimental settings. On the contrary, IκBαSR expression resulted in a prominent downregulation of BECN1 protein levels (Fig. 4D, right side, and E).
p65 positively modulates autophagy.
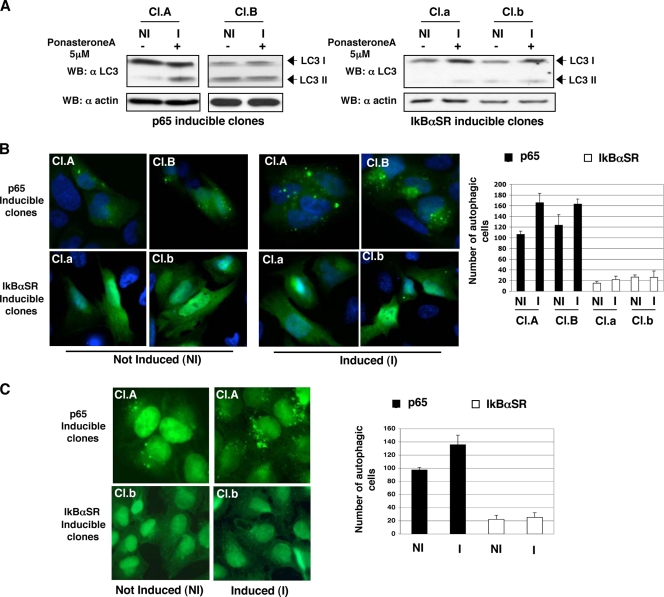
Since BECN1 is an essential autophagic protein (43), the possibility that p65 could regulate autophagy was investigated. As a first approach, the effects of p65- and IκBαSR-induced expression on endogenous LC3 processing were analyzed by means of immunoblotting. Indeed, following induction of autophagy, LC3 undergoes a regulated modification process that leads to the formation of a lipidated LC3 form termed LC3II. Lipidated LC3 is delivered to the autophagosome membranes, and this appears as a faster-migrating band when analyzed by Western blot assay. As shown in Fig. 5A (left side), LC3I processing efficiently occurred in p65-expressing clones but was strongly impaired in IκBαSR-expressing clones (Fig. 5A, right side). It must be noted that while IκBαSR induction clearly hampers constitutive autophagosome formation in both of the clones shown, p65 induction upregulates LC3 constitutive processing in only one of the two clones described. The basal autophagic flux of Cl.B is probably already elevated, and therefore it is not further increased by p65-mediated BECN1 transcription induction. Next, the effect of p65 and IκBαSR overexpression on green fluorescent protein (GFP)-LC3 body induction was evaluated by means of fluorescence microscopy. Similar to what emerged from Western blot analysis of the clones, induction of p65 expression clearly enhanced the number of GFP-LC3 bodies (Fig. 5B, top), while IκBαSR was almost completely unable to trigger GFP-LC3 body formation (Fig. 5B, bottom). Similar results were obtained by endogenous LC3 relocalization onto autophagosome membranes as a readout (Fig. 5C). Altogether, the data presented suggest that p65 can positively regulate constitutive autophagy, possibly due to its ability to regulate BECN1 expression.
FIG. 5.
p65 regulation of autophagy. (A) Clones A and B, inducible for p65, and clones a and b, inducible for IκBαSR, were treated with 5 μM ponasterone A for 6 h. Processing of endogenous LC3 (LC3I to LC3II) in cell lysates was monitored by immunoblotting. (B, left side) U2OS derivative clones A and B, inducible for p65 expression, and clones a and b, inducible for IκBαSR expression, were transfected with a small amount (100 ng) of GFP-hLC3 for 6 h and then induced (I) with ponasterone A for 12 h or not induced (NI). GFP-hLC3 fluorescence was evaluated, and some representative images are shown. (B, right side) Two hundred cells were counted for each experiment, and the number of cells presenting punctate GFP-hLC3 staining is reported. Data represent the means of at least three independent experiments. (C, left side) U2OS-derivative clones A, inducible for p65 expression, and b, inducible for IκBαSR, were induced (I) with ponasterone A for 10 h or not induced (NI), and endogenous LC3 was immunostained. Some representative images are shown on the right. (C, right side) the number of autophagic cells is reported in the graph. In all cases, cells presenting more than five autophagosomes per cell were scored as autophagic. (D and E, left side) Depletion of p65 decreases endogenous LC3 processing. U2OS and HeLa cells were transfected with a p65-specific siRNA or a scrambled control (Ctrl) siRNA for 72 h and subsequently not treated (NT) or treated for 12 h with 25 μM ceramide (Cera) or 10 μM tamoxifen (Tam). Effect of p65 knockdown on LC3I-to-LC3II conversion was evaluated by immunoblotting. (D and E, right side) Quantification of LC3I and LC3II bands was performed with the ImageJ program, and the LC3II/LC3I ratios reported are representative of autophagic process activation. The bar graph shows the n-fold LC3 conjugation decrease that occurred upon p65 depletion. The values shown are the mean results of three independent experiments. (F) Depletion of p65 decreases endogenous LC3 processing. HeLa cells were transfected with a p65-specific siRNA, a BECN1 siRNA, or a scrambled control siRNA for 72 h and subsequently treated for 20 h with 25 μM ceramide (C25) or 100 μM resveratrol (R) in the presence or absence of 0.5 mM leupeptin. Effects of knockdown of both p65 and BECN1 on LC3 processing were evaluated by immunoblotting. (G) U2OS cells were transfected with a p65-specific siRNA or a scrambled control siRNA. At 72 h later, GFP-hLC3 was transfected, and 6 h later, the cells were either not treated or treated with 10 μM tamoxifen for other 12 h. Autofluorescence of GFP-hLC3 was evaluated; representative images are shown on the left. (Right side) Two hundred cells were counted for each experiment, and the number of cell presenting punctate GFP-hLC3 staining was determined. Data represent the means of at least three independent experiments. Cells presenting more than five autophagosomes per cell were scored as autophagic. p65 interacts with the BECN1 promoter and upregulates BECN1 protein expression in activated T cells. WB, Western blotting.
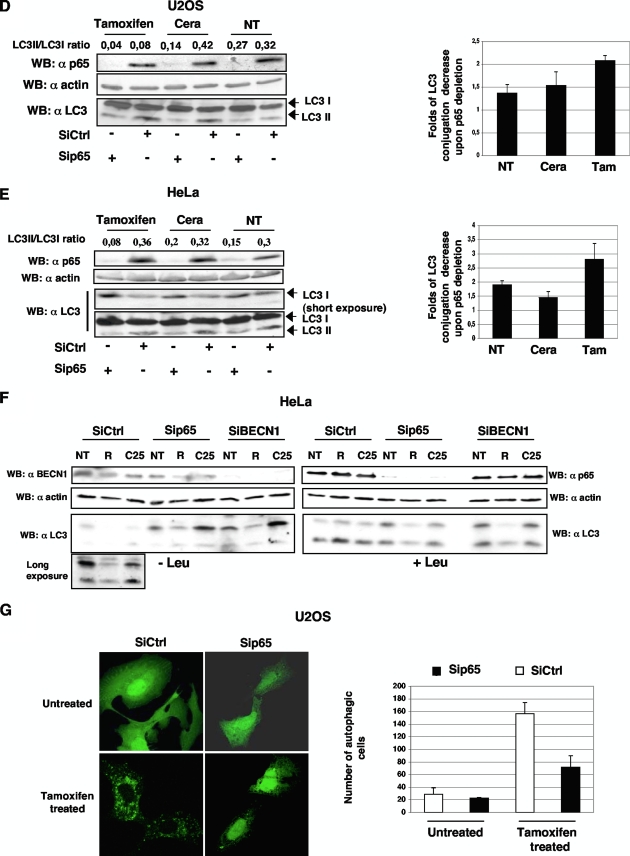
To further address this point, the role of endogenous p65 knockdown on the ability of U2OS and HeLa cells to undergo autophagy following either ceramide or tamoxifen treatment was analyzed. As shown in Fig. 5D and E, p65 depletion almost completely abolished LC3 processing, indicating that p65 could mediate autophagy induction in both U2OS and HeLa cells. Notably, p65 silencing also exerted the same kind of blockage in untreated control cells, again suggesting that p65 might affect cellular constitutive autophagy. We then asked whether the disappearance of the LC3II band following p65 silencing could be attributed to the induction of p65-mediated LC3 lysosomal processing. However, it is unlikely that such a process occurred, since a clear accumulation (more prominent in HeLa cells [Fig. 5E], shorter exposure of LC3 immunoblotting) of the unlipidated LC3 form was observed after p65 depletion. Indeed, pretreatment of the same cells with the lysosomal cathepsin inhibitor leupeptin confirmed that both constitutive autophagy and ceramide-induced autophagy were impaired following p65-specific knockdown (Fig. 5F). Notably, depletion of both BECN1 and p65 was completely unable to regulate resveratrol-induced noncanonical (BECN1-independent) autophagy (52) (Fig. 5F), further suggesting that p65 regulates autophagy through its ability to regulate BECN1 expression. Autophagosome formation was also monitored in tamoxifen-treated U2OS cells previously transfected with GFP-LC3 and a specific p65 siRNA or a scrambled control siRNA. Figure 5G shows that in the absence of any autophagic stimulus, GFP-LC3 staining appears diffuse both in the presence of p65 and after its depletion. Following tamoxifen treatment, GFP-LC3 clearly relocalizes into autophagosomal structures in cells transfected with the scrambled control siRNA, while p65-depleted cells displayed strongly impaired autophagosome formation. The ability of p65 to regulate autophagy was also tested in HEK293 cells overexpressing either p65 alone or the NF-κB inhibitors IκBαSR, p105, and p50. Again, in these settings, autophagy induced by ceramide occurred in p65-overexpressing cells and was blocked following IκBαSR, p105, or p50 overexpression (data not shown).
p65 binds to the BECN1 promoter in T cells.
In order to definitively confirm that BECN1 is a transcriptional target of p65, EMSA and ChIP experiments were performed. Jurkat T cells were chosen as a model system since they represent a classical model system to study NF-κB signaling and recent evidence has demonstrated that autophagy is induced upon T-cell receptor (TCR) activation (45). Intriguingly, TCR-mediated regulation of autophagy was shown to be required for proper T-cell homeostasis regulation. Similarly, p65 was also demonstrated to be engaged following TCR activation, contributing to T-cell homeostasis control (17).
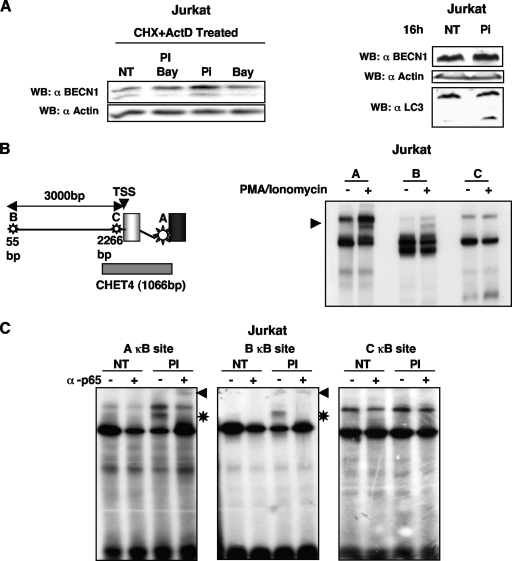
To assess the suitability of the system, Jurkat cells were pretreated with both actinomycin D and cycloheximide to block transcription/translation and overcome the interference of any IκBα-dependent feedback loop. After extensive washes to release the block, the cells were stimulated with PMA-ionomycin alone or together with the NF-κB inhibitor BAY 11-7082 and BECN1 levels were evaluated by immunoblotting. Upregulation of BECN1 protein expression was observed (Fig. 6A, left side). Notably, prolonged PMA-ionomycin treatment of Jurkat cells resulted in increased LC3 processing (Fig. 6A, right side).
FIG. 6.
p65 binds to the BECN1 promoter. (A, left side) Jurkat T cells were treated with cycloheximide (CHX, 5 μg/ml) and actinomycin D (ActD, 5 nM) for 10 h. Cells were washed once in order to remove the cycloheximide and actinomycin D and stimulated with PMA (200 ng/ml) plus ionomycin (300 ng/ml) or BAY 11-7082 (2.5 μM) or left untreated for 10 h. (A, right side) Jurkat cells were treated with PMA (200 ng/ml)-ionomycin (300 ng/ml) (PI) for 16 h, and LC3I-to-LC3 II conversion was monitored by immunoblotting. (B, right side) Nuclear extracts were prepared from Jurkat cells treated with PMA (200 ng/ml) plus ionomycin (300 ng/ml) for 2 h and from untreated Jurkat cells. Equal amounts of proteins were used for EMSA. Three κB sites, A, B, and C, predicted by TRANSFAC are schematically represented on the left. Probes encompassing all of the predicted κB sites were 32P labeled and used in the assays. The positions of the p65-DNA complexes are indicated by the arrowhead. TSS, transcription start site. (C) Nuclear extracts from PMA-ionomycin-treated Jurkat cells were preincubated in binding buffer or with an anti-p65 antibody and then incubated with the A, B, and C labeled probes. Asterisks mark the PMA-ionomycin-inducible complexes. The positions of the p65-DNA supershifted complex are indicated by the arrowheads. (D, left side) A ChIP assay reveals in vivo binding of endogenous p65 to the κB site predicted by CONFAC (A κB site). Jurkat cells were stimulated with PMA (200 ng/ml) plus ionomycin (300 ng/ml) (PI) for 5 h or left untreated, and protein-DNA complexes were cross-linked and immunoprecipitated (IP) with a p65-specific antibody (lanes 65) or an unrelated antibody (lanes nr). DNA was recovered and used as a template for PCR. Primers for a κB site inside the IκBα promoter and or for the β-actin promoter were used as positive and negative controls, respectively. (D, right side) ChIP was performed in triplicate as described above. Quantification of ChIPs by real-time PCR was performed to determine the amount of p65 binding to the indicated promoters compared with that of the input DNAs. Data are representative of three independent experiments. (E) Primary T lymphocytes (106) extracted from LNs of healthy C57BL/6 mice were left untreated or stimulated for 16 h with 1 μg/ml plate-bound anti-CD3, with anti-CD3 in combination with BAY 11-7082 (2.5 μM), or with BAY 11-7082 alone. BECN1 protein expression levels were monitored by Western blot (WB) analysis. Data are representative of three independent experiments. (F) Primary T lymphocytes (106) extracted from LNs of healthy C57BL/6 mice were treatment with PMA (10 ng/ml) plus ionomycin (300 ng/ml) alone or in combination with BAY 11-7082 (2.5 μM) or with BAY 11-7082 alone. Levels of BECN1 protein and LC3 processing were evaluated by immunoblotting. Data are representative of three independent experiments. (G) Primary CD8+ T-lymphocyte activation was monitored by FACS analysis. (Right side) CD-69 upregulation was evaluated following treatment with PMA (10 ng/ml) plus ionomycin (300 ng/ml) alone or in combination with BAY 11-7082 (2.5 μM) or with BAY 11-7082 alone. (Left side) Quantification of CD8+ T-lymphocyte activation. NT, not treated; Iono, ionomycin.
Nuclear extracts were prepared from PMA-ionomycin-treated and untreated Jurkat cells, and EMSAs were performed with three different 21-bp probes (A, B, and C, schematically represented in Fig. 6B, left side), each one encompassing the respective κB site predicted by TRANSFAC analysis. As shown in Fig. 6B (right side), in accordance with the luciferase data, the κB site located inside the first intron and conserved between humans and mice (A κB) showed the strongest binding activity and was clearly supershifted by an anti-p65 antibody. Competition assays confirmed the ability of p65 to specifically bind the A κB site (data not shown). Interestingly, another κB site located in a more upstream region of human BECN1 (B κB, Fig. 6B, left side) also gave rise to a retarded band that could be supershifted by an anti-p65 antibody (Fig. 6C), while the third predicted κB site (C κB site, Fig. 6B, left side) was unable to interact with any PMA-induced nuclear protein.
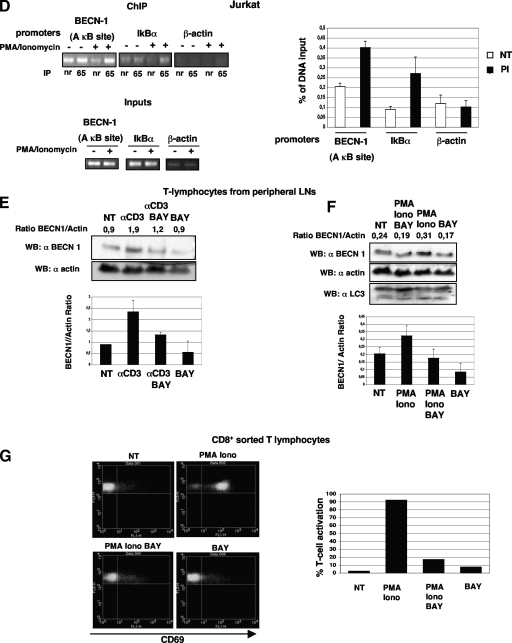
To investigate the interaction of p65 with the strongest κB site, ChIP assays were performed. Figure 6D shows that following PMA-ionomycin treatment, a promoter DNA sequence was amplified from the anti-p65 NF-κB immunoprecipitates with the specific PCR primers flanking the A κB site located inside the first intron of the BECN1 gene and conserved between humans and mice. Altogether, the data presented here demonstrate that BECN1 is a novel p65 target gene and suggest that following p65 activation in T cells, the subsequent regulation of BECN1 expression and autophagy could contribute to the control of T-cell homeostasis.
To explore whether the observed p65-driven increase in BECN1 levels was physiologically relevant, murine peripheral T lymphocytes were used to monitor BECN1 protein levels following TCR activation. Murine T lymphocytes from lymph nodes (LNs) of healthy C57BL/6 mice were activated with pharmacological agents (PMA-ionomycin) or with immobilized anti-CD3 (17). Notably, PMA in combination with ionomycin and anti-CD3 was also previously demonstrated to activate classical NF-κB signaling in T cells (17). We found that BECN1 protein levels increased after activation with immobilized anti-CD3 (Fig. 6E).
Stimulation of T lymphocytes with PMA in combination with ionomycin also resulted in a significant increase in BECN1protein levels (Fig. 6F). Interestingly, treatment with BAY-7085, a potent NF-κB inhibitor, led to a substantial decrease in BECN1 protein in murine T lymphocytes stimulated with either anti-CD3 (Fig. 6E) or PMA plus ionomycin (Fig. 6F). Indeed, treatment with the NF-κB inhibitor alone was sufficient to downregulate basal BECN1 protein levels. Notably, the ability of anti-CD3 to induce autophagy in primary T cells has already been demonstrated (45). In our experimental settings, treatment of healthy murine T cells with PMA in combination with ionomycin also resulted in an increase in LC3 processing (Fig. 6F).
Proper activation of murine T lymphocytes following PMA-ionomycin treatment was checked by monitoring the expression of the early activation marker CD-69 in sorted CD8+ lymphocytes. CD-69 exposure on the cell surface occurred after 3 h of PMA-ionomycin treatment (Fig. 6G). Importantly, stimulation of T lymphocytes with PMA-ionomycin in combination with BAY-7085 significantly blocked T-cell activation. Altogether, these results argue for a role for p65-mediated induction of BECN1 in T-cell activation and suggest that this regulation might be important for autophagy induction following TCR engagement.
DISCUSSION
The NF-κB family of transcription factors gives rise to a broad and sometimes contradictory spectrum of biological responses, depending on the cellular context and on the inducing stimulus. In particular, NF-κB transcription factors have been shown to either promote or inhibit cell death (19). Indeed, NF-κB family member p65 has been shown to be required to induce p53-dependent apoptosis following hydrogen peroxide, deferoxamine mesylate, or adriamycin treatment (49). On the other hand, p65 efficiently counteracts TNF-α-induced apoptosis (49) through its ability to directly induce the expression of several antiapoptotic genes, including those for inhibitors of apoptosis, Bcl2, and BclXL.
In this work, we highlighted the relevance of NF-κB family member p65 to the control of autophagy, an important cellular mechanism which functions, similarly to NF-κB signaling, in promoting both cell death and cell survival, depending on the specific cellular context and on the cellular genetic background. Autophagy induces cell demise in tamoxifen- and resveratrol-treated MCF-7 cells, and its inhibition by 3-methyladenine prevents cell death (41, 51, 52). Other treatments such as ionizing radiation induce autophagy in several tumor cell lines, but the exact outcome of autophagy induction in these settings remains to be clarified. Inhibition of autophagy in radiation-treated cells increases cytotoxicity, pointing to a prosurvival role for autophagy (42). Similarly, autophagy works as a strategy to survive exposure to alkylating agents and TNF-related apoptosis-inducing ligand chemotherapeutic treatment (2, 22) and counteracts metabolic stress occurring in the inner core of solid tumors (12). Notably, NF-κB has also been frequently implicated in chemoresistance (3).
Recently, we addressed the role of calpain in the modulation of cell death, highlighting the importance of this enzyme in inducing cytoprotective autophagy in response to different stimuli, including ceramide, rapamycin, and etoposide (13). Notably, impairment of autophagy by calpain depletion was more severe than that caused by ATG5 depletion, indicating that additional and probably upstream prosurvival mechanisms, such as NF-κB prosurvival activation, are lost in the absence of calpain. Indeed, calpain was previously demonstrated to be involved in NF-κB activation, contributing to its prosurvival functions in response to ceramide (14). Furthermore, several studies have demonstrated that ceramide and tamoxifen, besides triggering apoptosis, also induce autophagy (31, 51). Interestingly, many stimuli known to induce autophagy, such as reactive oxygen species, ceramide, TNF-α, and TNF-related apoptosis-inducing ligand, but also viral and bacterial infections, also act as NF-κB inducers (7, 14, 23, 24). Altogether, these pieces of evidence may suggest the involvement of NF-κB in autophagy induction. In this work, we clearly show such an involvement, demonstrating that p65 binds to the promoter of the essential autophagic gene BECN1 and regulates its expression in response to ceramide and tamoxifen treatment. p65 activation increases BECN1 expression and, as a result, positively regulates autophagy. Importantly, in our experimental settings, we observed that BECN1 expression and autophagy were regulated by p65 both under basal conditions and following the application of some autophagic stimuli. Interestingly, a very recent work highlighted the existence of another form of autophagy induced by resveratrol treatment in some cell lines, referred to as noncanonical autophagy, that does not require BECN1 (52). In accordance with this study, we found that p65 was completely unable to regulate resveratrol-induced noncanonical autophagy in HeLa cells, confirming that only BECN1-mediated routes of autophagy induction can be regulated by p65. These findings strengthen the relationship between p65 and canonical, BECN1-mediated autophagy, suggesting that p65 might be required for constitutive canonical autophagy, which promotes organelle and protein quality control, maintaining cellular homeostasis. Consistently, NF-κB has emerged as a key transcription factor required for the proper development of immune system cells and required for homeostasis control in several tissues. Intriguingly, similarly to p65, autophagy has recently been demonstrated to be critical for T-cell homeostasis maintenance (17, 45). Autophagy is required for the survival of interleukin-3-dependent hematopoietic cell lines following cytokine withdrawal (37) and ATG5−/− T-cell survival and proliferation (45). Strikingly, the phenotype of ATG5−/− T-cell chimeras closely resembles that observed in p65−/− chimeras. Overlapping features of these models are the diminished proliferative response to TCR stimulation with anti-CD3 and PMA-ionomycin and the reduced number of cells in the thymuses of mice with transplanted RelA−/− or ATG5−/− fetal liver cells (17, 45). Autophagy has been shown to occur in primary T lymphocytes following TCR activation induced by anti-CD3 or PMA-ionomycin treatment (35, 45). Since p65 is required for TCR signaling, it is possible that p65-mediated induction of BECN1 expression could be important for autophagy induction in these settings. Consistent with this hypothesis, our experiments demonstrated that inhibition of p65 activation impaired T-cell activation and BECN1 upregulation, suggesting that p65 could contribute to the homeostasis control of T lymphocytes. Notably, protein kinase C θ, which is a key player in TCR signaling and is specifically involved in NF-κB activation (21), has recently been implicated in the regulation of ER stress-induced autophagy (50). Intriguingly, many ER stress stimuli, such as tunicamycin and thapsigargin, besides activating protein kinase C θ, also induce eIF2α phosphorylation, which has been shown to activate NF-κB (15).
In contrast to our findings, highlighting a positive role for p65 classical signaling in the regulation of autophagy, it has been reported that NF-κB activation represses TNF-α-induced autophagy (16). This discrepancy can be explained by the fact that both autophagy and NF-κB signaling are context and stimulus dependent. Indeed, in accordance with this observation, the authors of the above paper reported that autophagy occurs in the same manner in both NF-κB-competent and NF-κB-incompetent cell lines following nutrient starvation. Interestingly, in line with our results, it is shown that inhibition of p65 does not lead to an increase in BECN1 transcription following TNF-α treatment but results in an increase in BECN1 protein, suggesting a posttranslational regulation of this protein. In the same work, depletion of p65 in MCF-7 cells was sufficient for autophagy induction (16). In agreement with these findings, we observed that p65 overexpression could not activate BECN1 promoter in some luciferase reporter assays performed with the same cell line (data not shown). One possible explanation is that the p65 response is strictly cell context dependent. Evidence exists that in this ER-positive and selective estrogen modulator (one of which is tamoxifen)-sensitive cell lines, NF-κB signaling should be attributed to p50 and Bcl3 rather than to p65 (68, 69). Another interesting work demonstrated that autophagy represents a new route for NF-κB-inducing kinase degradation and negatively regulates alternative NF-κB pathways (47). In light of these findings and of our data, it is tempting to speculate that autophagy, once acutely induced through a classical mode of NF-κB activation, could terminate the whole NF-κB pathway, working as a negative feedback mechanism.
Supplementary Material
Acknowledgments
We are grateful to F. Benvenuti (The International Centre for Genetic Engineering and Biotechnology [ICGEB]) for supplying murine T lymphocytes and for protocols and FACS analysis of T-cell activation. We thank M. Lusic (ICGEB) for providing ChIP protocol suggestions and M. Giacca (ICGEB) for access to the FACS apparatus and the sonicator. We thank S. Giacomel for technical support. We are grateful to I. Tanida for the pEGFP-hLC3 vector, to T. Yoshimori for the pEGFP-rLC3 vector, to N. Perkins for the pRSVhRelA and pRSVp50 vectors, to Shao-Cong Sun for the pCMV4HA-IKBαSR vector, and to G. Del Sal for the pCMVHA-E2F vector.
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and TRANSlational and Functional Onco-Genomics (TRANSFOG) (CEE no. 503438 to C.S.).
Footnotes
Published ahead of print on 16 March 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aita, V. M., X. H. Liang, V. V. Murty, D. L. Pincus, W. Yu, E. Cayanis, S. Kalachikov, T. C. Gilliam, and B. Levine. 1999. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 5959-65. [DOI] [PubMed] [Google Scholar]
- 2.Amaravadi, R. K., D. Yu, J. J. Lum, T. Bui, M. A. Christophorou, G. I. Evan, A. Thomas-Tikhonenko, and C. B. Thompson. 2007. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Investig. 117326-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlt, A., J. Vorndamm, S. Muerkoster, H. Yu, W. E. Schmidt, U. R. Folsch, and H. Schafer. 2002. Autocrine production of interleukin 1β confers constitutive nuclear factor κB activity and chemoresistance in pancreatic carcinoma cell lines. Cancer Res. 62910-916. [PubMed] [Google Scholar]
- 4.Bohensky, J., I. M. Shapiro, S. Leshinsky, S. P. Terkhorn, C. S. Adams, and V. Srinivas. 2007. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy 3207-214. [DOI] [PubMed] [Google Scholar]
- 5.Boland, M. P., and L. A. O'Neill. 1998. Ceramide activates NFκB by inducing the processing of p105. J. Biol. Chem. 27315494-15500. [DOI] [PubMed] [Google Scholar]
- 6.Boya, P., R. A. Gonzalez-Polo, N. Casares, J. L. Perfettini, P. Dessen, N. Larochette, D. Metivier, D. Meley, S. Souquere, T. Yoshimori, G. Pierron, P. Codogno, and G. Kroemer. 2005. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 251025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bubici, C., S. Papa, C. G. Pham, F. Zazzeroni, and G. Franzoso. 2006. The NF-κB-mediated control of ROS and JNK signaling. Histol. Histopathol. 2169-80. [DOI] [PubMed] [Google Scholar]
- 8.Colell, A., J. E. Ricci, S. Tait, S. Milasta, U. Maurer, L. Bouchier-Hayes, P. Fitzgerald, A. Guio-Carrion, N. J. Waterhouse, C. W. Li, B. Mari, P. Barbry, D. D. Newmeyer, H. M. Beere, and D. R. Green. 2007. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 129983-997. [DOI] [PubMed] [Google Scholar]
- 9.Crighton, D., S. Wilkinson, J. O'Prey, N. Syed, P. Smith, P. R. Harrison, M. Gasco, O. Garrone, T. Crook, and K. M. Ryan. 2006. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126121-134. [DOI] [PubMed] [Google Scholar]
- 10.Cuervo, A. M., W. Hu, B. Lim, and J. F. Dice. 1998. IκB is a substrate for a selective pathway of lysosomal proteolysis. Mol. Biol. Cell 91995-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel, F., A. Legrand, D. Pessayre, N. Vadrot, V. Descatoire, and D. Bernuau. 2006. Partial Beclin 1 silencing aggravates doxorubicin- and Fas-induced apoptosis in HepG2 cells. World J. Gastroenterol. 122895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degenhardt, K., R. Mathew, B. Beaudoin, K. Bray, D. Anderson, G. Chen, C. Mukherjee, Y. Shi, C. Gelinas, Y. Fan, D. A. Nelson, S. Jin, and E. White. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 1051-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demarchi, F., C. Bertoli, T. Copetti, I. Tanida, C. Brancolini, E. L. Eskelinen, and C. Schneider. 2006. Calpain is required for macroautophagy in mammalian cells. J. Cell Biol. 175595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demarchi, F., C. Bertoli, P. A. Greer, and C. Schneider. 2005. Ceramide triggers an NF-κB-dependent survival pathway through calpain. Cell Death Differ. 12512-522. [DOI] [PubMed] [Google Scholar]
- 15.Deng, J., P. D. Lu, Y. Zhang, D. Scheuner, R. J. Kaufman, N. Sonenberg, H. P. Harding, and D. Ron. 2004. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol. Cell. Biol. 2410161-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djavaheri-Mergny, M., M. Amelotti, J. Mathieu, F. Besancon, C. Bauvy, S. Souquere, G. Pierron, and P. Codogno. 2006. NF-κB activation represses tumor necrosis factor-α-induced autophagy. J. Biol. Chem. 28130373-30382. [DOI] [PubMed] [Google Scholar]
- 17.Doi, T. S., T. Takahashi, O. Taguchi, T. Azuma, and Y. Obata. 1997. NF-κB RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J. Exp. Med. 185953-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabre, C., G. Carvalho, E. Tasdemir, T. Braun, L. Ades, J. Grosjean, S. Boehrer, D. Metivier, S. Souquere, G. Pierron, P. Fenaux, and G. Kroemer. 2007. NF-κB inhibition sensitizes to starvation-induced cell death in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene 264071-4083. [DOI] [PubMed] [Google Scholar]
- 19.Foo, S. Y., and G. P. Nolan. 1999. NF-κB to the rescue: RELs, apoptosis and cellular transformation. Trends Genet. 15229-235. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, S., V. Tergaonkar, C. V. Rothlin, R. G. Correa, V. Bottero, P. Bist, I. M. Verma, and T. Hunter. 2006. Essential role of tuberous sclerosis genes TSC1 and TSC2 in NF-κB activation and cell survival. Cancer Cell 10215-226. [DOI] [PubMed] [Google Scholar]
- 21.Grumont, R., P. Lock, M. Mollinari, F. M. Shannon, A. Moore, and S. Gerondakis. 2004. The mitogen-induced increase in T cell size involves PKC and NFAT activation of Rel/NF-κB-dependent c-myc expression. Immunity 2119-30. [DOI] [PubMed] [Google Scholar]
- 22.Han, J., W. Hou, L. A. Goldstein, C. Lu, D. B. Stolz, X. M. Yin, and H. Rabinowich. 2008. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J. Biol. Chem. 28319665-19677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayden, M. S., A. P. West, and S. Ghosh. 2006. NF-κB and the immune response. Oncogene 256758-6780. [DOI] [PubMed] [Google Scholar]
- 24.Hiscott, J., T. L. Nguyen, M. Arguello, P. Nakhaei, and S. Paz. 2006. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene 256844-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbard, T. J., B. L. Aken, K. Beal, B. Ballester, M. Caccamo, Y. Chen, L. Clarke, G. Coates, F. Cunningham, T. Cutts, T. Down, S. C. Dyer, S. Fitzgerald, J. Fernandez-Banet, S. Graf, S. Haider, M. Hammond, J. Herrero, R. Holland, K. Howe, K. Howe, N. Johnson, A. Kahari, D. Keefe, F. Kokocinski, E. Kulesha, D. Lawson, I. Longden, C. Melsopp, K. Megy, P. Meidl, B. Ouverdin, A. Parker, A. Prlic, S. Rice, D. Rios, M. Schuster, I. Sealy, J. Severin, G. Slater, D. Smedley, G. Spudich, S. Trevanion, A. Vilella, J. Vogel, S. White, M. Wood, T. Cox, V. Curwen, R. Durbin, X. M. Fernandez-Suarez, P. Flicek, A. Kasprzyk, G. Proctor, S. Searle, J. Smith, A. Ureta-Vidal, and E. Birney. 2007. Ensembl 2007. Nucleic Acids Res. 35D610-D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung, J. H., I. J. Su, H. Y. Lei, H. C. Wang, W. C. Lin, W. T. Chang, W. Huang, W. C. Chang, Y. S. Chang, C. C. Chen, and M. D. Lai. 2004. Endoplasmic reticulum stress stimulates the expression of cyclooxygenase-2 through activation of NF-κB and pp38 mitogen-activated protein kinase. J. Biol. Chem. 27946384-46392. [DOI] [PubMed] [Google Scholar]
- 27.Jia, G., G. Cheng, D. M. Gangahar, and D. K. Agrawal. 2006. Insulin-like growth factor-1 and TNF-α regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol. Cell Biol. 84448-454. [DOI] [PubMed] [Google Scholar]
- 28.Karanam, S., and C. S. Moreno. 2004. CONFAC: automated application of comparative genomic promoter analysis to DNA microarray datasets. Nucleic Acids Res. 32W475-W484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuma, A., M. Hatano, M. Matsui, A. Yamamoto, H. Nakaya, T. Yoshimori, Y. Ohsumi, T. Tokuhisa, and N. Mizushima. 2004. The role of autophagy during the early neonatal starvation period. Nature 4321032-1036. [DOI] [PubMed] [Google Scholar]
- 30.Kuma, A., N. Mizushima, N. Ishihara, and Y. Ohsumi. 2002. Formation of the approximately 350-kDa Apg12-Apg5. Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 27718619-18625. [DOI] [PubMed] [Google Scholar]
- 31.Lavieu, G., F. Scarlatti, G. Sala, S. Carpentier, T. Levade, R. Ghidoni, J. Botti, and P. Codogno. 2006. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J. Biol. Chem. 2818518-8527. [DOI] [PubMed] [Google Scholar]
- 32.Levine, B., and J. Abrams. 2008. p53: The Janus of autophagy? Nat. Cell Biol. 10637-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine, B., and G. Kroemer. 2008. Autophagy in the pathogenesis of disease. Cell 13227-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine, B., and J. Yuan. 2005. Autophagy in cell death: an innocent convict? J. Clin. Investig. 1152679-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, C., E. Capan, Y. Zhao, J. Zhao, D. Stolz, S. C. Watkins, S. Jin, and B. Lu. 2006. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J. Immunol. 1775163-5168. [DOI] [PubMed] [Google Scholar]
- 36.Lukiw, W. J., A. Ottlecz, G. Lambrou, M. Grueninger, J. Finley, H. W. Thompson, and N. G. Bazan. 2003. Coordinate activation of HIF-1 and NF-κB DNA binding and COX-2 and VEGF expression in retinal cells by hypoxia. Investig. Ophthalmol. Vis. Sci. 444163-4170. [DOI] [PubMed] [Google Scholar]
- 37.Lum, J. J., D. E. Bauer, M. Kong, M. H. Harris, C. Li, T. Lindsten, and C. B. Thompson. 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120237-248. [DOI] [PubMed] [Google Scholar]
- 38.Matys, V., O. V. Kel-Margoulis, E. Fricke, I. Liebich, S. Land, A. Barre-Dirrie, I. Reuter, D. Chekmenev, M. Krull, K. Hornischer, N. Voss, P. Stegmaier, B. Lewicki-Potapov, H. Saxel, A. E. Kel, and E. Wingender. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34D108-D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizushima, N., A. Kuma, Y. Kobayashi, A. Yamamoto, M. Matsubae, T. Takao, T. Natsume, Y. Ohsumi, and T. Yoshimori. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 1161679-1688. [DOI] [PubMed] [Google Scholar]
- 40.Monks, N. R., and A. B. Pardee. 2006. Targeting the NF-κB pathway in estrogen receptor negative MDA-MB-231 breast cancer cells using small inhibitory RNAs. J. Cell. Biochem. 98221-233. [DOI] [PubMed] [Google Scholar]
- 41.Opipari, A. W., Jr., L. Tan, A. E. Boitano, D. R. Sorenson, A. Aurora, and J. R. Liu. 2004. Resveratrol-induced autophagocytosis in ovarian cancer cells. Cancer Res. 64696-703. [DOI] [PubMed] [Google Scholar]
- 42.Paglin, S., T. Hollister, T. Delohery, N. Hackett, M. McMahill, E. Sphicas, D. Domingo, and J. Yahalom. 2001. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 61439-444. [PubMed] [Google Scholar]
- 43.Pattingre, S., L. Espert, M. Biard-Piechaczyk, and P. Codogno. 2008. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie 90313-323. [DOI] [PubMed] [Google Scholar]
- 44.Polager, S., M. Ofir, and D. Ginsberg. 2008. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 274860-4864. [DOI] [PubMed] [Google Scholar]
- 45.Pua, H. H., I. Dzhagalov, M. Chuck, N. Mizushima, and Y. W. He. 2007. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 20425-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pulecio, J., E. Tagliani, A. Scholer, F. Prete, L. Fetler, O. R. Burrone, and F. Benvenuti. 2008. Expression of Wiskott-Aldrich syndrome protein in dendritic cells regulates synapse formation and activation of naive CD8+ T cells. J. Immunol. 1811135-1142. [DOI] [PubMed] [Google Scholar]
- 47.Qing, G., P. Yan, Z. Qu, H. Liu, and G. Xiao. 2007. Hsp90 regulates processing of NF-κB2 p100 involving protection of NF-κB-inducing kinase (NIK) from autophagy-mediated degradation. Cell Res. 17520-530. [DOI] [PubMed] [Google Scholar]
- 48.Qu, X., J. Yu, G. Bhagat, N. Furuya, H. Hibshoosh, A. Troxel, J. Rosen, E. L. Eskelinen, N. Mizushima, Y. Ohsumi, G. Cattoretti, and B. Levine. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 1121809-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan, K. M., J. O'Prey, and K. H. Vousden. 2004. Loss of nuclear factor-κB is tumor promoting but does not substitute for loss of p53. Cancer Res. 644415-4418. [DOI] [PubMed] [Google Scholar]
- 50.Sakaki, K., and R. J. Kaufman. 2008. Regulation of ER stress-induced macroautophagy by protein kinase C. Autophagy 4841-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scarlatti, F., C. Bauvy, A. Ventruti, G. Sala, F. Cluzeaud, A. Vandewalle, R. Ghidoni, and P. Codogno. 2004. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J. Biol. Chem. 27918384-18391. [DOI] [PubMed] [Google Scholar]
- 52.Scarlatti, F., R. Maffei, I. Beau, P. Codogno, and R. Ghidoni. 2008. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 151318-1329. [DOI] [PubMed] [Google Scholar]
- 53.Sha, W. C., H. C. Liou, E. I. Tuomanen, and D. Baltimore. 1995. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell 80321-330. [DOI] [PubMed] [Google Scholar]
- 54.Sivaprasad, U., and A. Basu. 2008. Inhibition of ERK attenuates autophagy and potentiates tumor necrosis factor-α-induced cell death in MCF-7 cells. J. Cell. Mol. Med. 121265-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takacs-Vellai, K., T. Vellai, A. Puoti, M. Passannante, C. Wicky, A. Streit, A. L. Kovacs, and F. Muller. 2005. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr. Biol. 151513-1517. [DOI] [PubMed] [Google Scholar]
- 56.Tanida, I., T. Ueno, and E. Kominami. 2004. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J. Biol. Chem. 27947704-47710. [DOI] [PubMed] [Google Scholar]
- 57.Tasdemir, E., M. C. Maiuri, L. Galluzzi, I. Vitale, M. Djavaheri-Mergny, M. D'Amelio, A. Criollo, E. Morselli, C. Zhu, F. Harper, U. Nannmark, C. Samara, P. Pinton, J. M. Vicencio, R. Carnuccio, U. M. Moll, F. Madeo, P. Paterlini-Brechot, R. Rizzuto, G. Szabadkai, G. Pierron, K. Blomgren, N. Tavernarakis, P. Codogno, F. Cecconi, and G. Kroemer. 2008. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 10676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tracy, K., B. C. Dibling, B. T. Spike, J. R. Knabb, P. Schumacker, and K. F. Macleod. 2007. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell. Biol. 276229-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333169-174. [DOI] [PubMed] [Google Scholar]
- 60.van Uden, P., N. S. Kenneth, and S. Rocha. 2008. Regulation of hypoxia-inducible factor-1α by NF-κB. Biochem. J. 412477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 216820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weisz, L., A. Damalas, M. Liontos, P. Karakaidos, G. Fontemaggi, R. Maor-Aloni, M. Kalis, M. Levrero, S. Strano, V. G. Gorgoulis, V. Rotter, G. Blandino, and M. Oren. 2007. Mutant p53 enhances nuclear factor κB activation by tumor necrosis factor α in cancer cells. Cancer Res. 672396-2401. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto, Y., U. N. Verma, S. Prajapati, Y. T. Kwak, and R. B. Gaynor. 2003. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature 423655-659. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, H., M. Bosch-Marce, L. A. Shimoda, Y. S. Tan, J. H. Baek, J. B. Wesley, F. J. Gonzalez, and G. L. Semenza. 2008. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 28310892-10903. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Zhang, J. Y., C. L. Green, S. Tao, and P. A. Khavari. 2004. NF-κB RelA opposes epidermal proliferation driven by TNFR1 and JNK. Genes Dev. 1817-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, J., J. J. Brault, A. Schild, P. Cao, M. Sandri, S. Schiaffino, S. H. Lecker, and A. L. Goldberg. 2007. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6472-483. [DOI] [PubMed] [Google Scholar]
- 67.Zheng, Y., F. Ouaaz, P. Bruzzo, V. Singh, S. Gerondakis, and A. A. Beg. 2001. NF-κ B RelA (p65) is essential for TNF-α-induced fas expression but dispensable for both TCR-induced expression and activation-induced cell death. J. Immunol. 1664949-4957. [DOI] [PubMed] [Google Scholar]
- 68.Zhou, Y., S. Eppenberger-Castori, U. Eppenberger, and C. C. Benz. 2005. The NFκB pathway and endocrine-resistant breast cancer. Endocr. Relat. Cancer 12(Suppl. 1)S37-S46. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, Y., C. Yau, J. W. Gray, K. Chew, S. H. Dairkee, D. H. Moore, U. Eppenberger, S. Eppenberger-Castori, and C. C. Benz. 2007. Enhanced NFκB and AP-1 transcriptional activity associated with antiestrogen resistant breast cancer. BMC Cancer 759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.