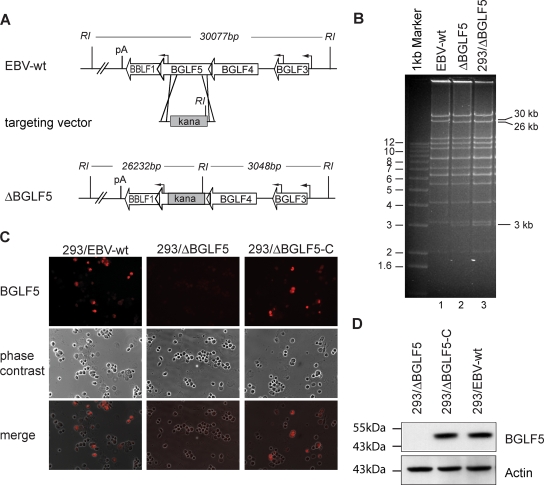
FIG. 1.
Construction of a ΔBGLF5 recombinant virus. (A) Schematic map of the EBV genome segment that encompasses the BGLF5 gene in EBV-wt and after homologous recombination with the targeting vector carrying the kanamycin resistance gene. The overlapping BGLF4 and BBLF1 coding regions are not affected by the recombination. The cleavage sites for EcoRI (RI) and the expected fragment sizes for cleavage of EBV-wt and ΔBGLF5 genomes are given. pA, polyadenylation site, kana, kanamycin. (B) EcoRI restriction fragment analysis of EBV-wt (lane 1) and ΔBGLF5 mutant genomes (lane 2) after construction in E. coli or after rescue from stably transfected 293 cells (293/ΔBGLF5) (lane 3). The result is consistent with the predicted restriction pattern (see panel A). (C) Immunofluorescence analysis of lytically induced 293/ΔBGLF5, 293/EBV-wt and transcomplemented 293/ΔBGLF5 cells (293/ΔBGLF5-C) with a BGLF5-specific antibody. No BGLF5 protein was detected in 293/ΔBGLF5 mutant cells 2 days after induction. (D) Western blot analysis on extracts from induced 293/EBV-wt, 293/ΔBGLF5, and 293/ΔBGLF5-C cells, 48 h after induction. The transcomplemented 293/ΔBGLF5-C and 293/EBV-wt producer lines express BGLF5 at comparable levels, 293/ΔBGLF5 cells were BGLF5 negative. Staining of the same blot with an actin-specific antibody served as a loading control.