Abstract
Previously we showed that the E1A binding proteins p300 and CBP negatively regulate c-Myc in quiescent cells and that binding of E1A to p300 results in the induction of c-Myc and thereby induction of S phase. We demonstrated that p300 and HDAC3 cooperate with the transcription factor YY1 at an upstream YY1 binding site and repress the Myc promoter. Here we show that the small E1A protein induces c-Myc by interfering with the protein-protein interaction between p300, YY1, and HDAC3. Wild-type E1A but not the E1A mutants that do not bind to p300 interfered in recruitment of YY1, p300, and HDAC3 to the YY1 binding site. As E1A started to accumulate after infection, it transiently associated with promoter-bound p300. Subsequently, YY1, p300, and HDAC3 began to dissociate from the promoter. Later in infection, E1A dissociated from the promoter as well as p300, YY1, and HDAC3. Removal of HDAC3 from the promoter correlated with increased acetylation of Myc chromatin and induction. In vivo E1A stably associated with p300 and dissociated YY1 and HDAC3 from the trimolecular complex. In vitro protein-protein interaction studies indicated that E1A initially binds to the p300-YY1-HDAC3 complex, briefly associates with it, and then dissociates the complex, recapitulating somewhat the in vivo situation. Thus, E1A binding to the C-terminal region of p300 disrupts the important corepressor function provided by p300 in repressing c-Myc. Our results reveal a novel mechanism by which a viral oncoprotein activates c-Myc in quiescent cells and raise the possibility that the oncoproteins encoded by the small-DNA tumor viruses may use this mechanism to induce c-Myc, which may be critical for cell transformation.
Cell transformation and induction of DNA synthesis in quiescent cells by the adenovirus (Ad) transforming protein E1A are dependent on its binding to and modifying the activities of several host proteins, including p400, p300/CBP, and the pocket family proteins pRb, p107, and p130 (3, 9, 10, 25, 30). Several of these proteins associate with cellular repressor complexes and inhibit transcription factors involved in the induction of cell cycle S phase (22, 23, 30). The E1A binding proteins p300 and CBP are two nuclear phosphoproteins that coactivate a large number of transcription factors to stimulate transcription. They also contain intrinsic histone acetyltransferase activity that acetylates chromatin and thereby decondenses it to facilitate transcription (13).
In quiescent cells, binding of E1A to p300 is essential for the induction of DNA synthesis and cell transformation (25, 27, 33). For the past several years, we have been investigating the role of p300/CBP in quiescent cells and the cell cycle G1/S transition and the consequences of binding of E1A to p300 in the induction of S phase. We showed that both p300 and CBP negatively regulate the transition of cells from G0/G1 to S phase by keeping c-Myc in a repressed state and that normal amounts of both of these coactivators are essential for repressing c-Myc (1, 18, 29). Further, we showed that wild-type (WT) E1A, but not the E1A mutants that do not bind to p300, induces S phase by inducing c-Myc (2, 18). In a more recent report, we showed that the C-terminal region of p300 provides a corepressor function in repressing c-Myc (30). The transcription factor YY1 binds to an upstream YY1 binding site of the Myc promoter and recruits p300 and HDAC3. HDAC3 thus recruited to the YY1-p300 complex deacetylates Myc chromatin and represses transcription. The repressive activity of p300 is independent of the intrinsic histone acetyltransferase (HAT) activity of p300 (1). Sumoylation of p300 also is not necessary for the repression, since p300 in which the two sumoylation sites were mutated was found to be as efficient as WT p300 in repressing c-Myc (30). Furthermore, we recently showed that simian virus 40 (SV40) large T also has a capacity to relieve the repression of c-Myc by p300 (31), raising the possibility that deregulation of Myc by the DNA tumor virus T antigens may be an essential prerequisite for cell transformation.
c-Myc plays a pivotal role in a number of pathways that control cell growth and differentiation, and deregulation of c-Myc is associated with several forms of human cancers (5, 6). In this work, we studied the mechanism by which E1A relieves the repression of c-Myc by p300 in quiescent cells. We showed that the transforming E1A protein interferes with the recruitment of YY1, p300, and HDAC3 to the upstream YY1 binding site of the Myc promoter and also disrupts the interaction between these three proteins. E1A interferes with the protein-protein interactions among these transcriptional effectors both in vivo and in vitro. E1A interference with p300 function is dependent on E1A binding to p300 through its third cysteine-histidine-rich (CH3) region. Early after infection, E1A transiently associated with p300 bound to the chromatin, resulting in the dissociation of p300, YY1, and HDAC3 from the promoter. At later times, neither E1A nor other transcriptional effectors referred to above associated with chromatin. Our results suggest that E1A initially binds to p300 at the chromatin level and causes the dissociation of p300, YY1, and HDAC3 from the chromatin, leading to the relief of repression. E1A is also dissociated from chromatin in this process. These results provide new insights into the mechanism by which a virus-encoded oncoprotein induces c-Myc.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
MCF10A cells are immortalized but nontransformed human breast epithelial cells (32). SaOS2 cells are osteosarcoma cells, and rat12 cells are a derivative of rat1 cells and have similar growth properties. Further details and growth conditions of these cell lines can be obtained from our previous reports (1, 18). Adb-gal is an Ad vector expressing the beta-galactosidase gene from the E1A region (18). Adp300AT2 is an Ad vector expressing a FLAG-tagged HAT-defective p300 mutant that contains six amino acid substitutions in its HAT domain (p300AT2) (19). The Ad mutant expressing only the 243-amino-acid (aa) transforming E1A protein (dl1500) (24) was used as the WT virus in these studies. The mutant dl2-36 contains a deletion of 36 aa from residue 2 to 36, and the mutant RG2 has an arginine-to-glycine change in the second codon (34). The plasmid del6 is a truncated human c-Myc promoter-reporter plasmid derived from a larger Myc promoter-luciferase reporter plasmid (15) and contains 640 bp of the upstream promoter sequence (1). Plasmids expressing hemagglutinin-tagged p300 and p300delE1A were gifts of D. Livingston (Harvard University). The plasmid p300delE1A contains a 132-aa deletion (aa 1739 to 1871) encompassing the E1A binding region (7).
Promoter-reporter assays.
Rat12 cells (4 × 105 to 6 × 105 per well) were seeded in six-well plates overnight and then transfected with the Myc promoter-reporter plasmid del6 along with various expression plasmids, as indicated in figure legends, using Lipofectamine 2000 (Invitrogen). The total amount of DNA for each transfection was kept constant by adding empty vectors as appropriate. Transfected cells were maintained overnight in growth medium and then serum starved for 36 h, harvested, and lysed in a lysis buffer (Promega). The luciferase activity in the lysates was quantified using equal quantities of protein as described previously (29). Renilla luciferase was used for transfection efficiency (Promega). In our assay, E1A did not affect Renilla luciferase activity by sequestering p300.
In vivo and in vitro protein-protein interaction studies.
For in vivo protein-protein interaction studies, serum-starved MCF10A cells were infected with Ad variants expressing WT E1A, RG2, or dl2-36 as appropriate. At 16 h following infection, cells were collected, lysed in a lysis buffer consisting of 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, and protease inhibitors on ice for 20 min. The cell lysates were passed through a 21-gauge hypodermic needle and clarified by centrifugation at 12,000 rpm in a Sorvall RC5B centrifuge for 30 min at 4°C. Cell lysates equivalent to 2 mg protein were immunoprecipitated with antibodies as appropriate, and the immunoprecipitated proteins were identified by Western immunoblotting using the appropriate antibodies (indicated in legends to figures). For in vitro glutathione S-transferase (GST) pull-down assays, GST fusion proteins containing YY1, HDAC3, p300CT (aa 1572 to 2370), WTE1A, and the E1A mutants dl2-36 and RG2 were prepared in Escherichia coli using standard procedures (30). Protein binding assays were carried out on ice for the indicated periods using HEPES buffer (20 mM, pH 7.9) containing KCl (150 mM), EDTA (0.2 mM), and complete protease inhibitor cocktail. The reaction was terminated by diluting the sample 10-fold with the lysis buffer described above. Following incubation, the proteins were immunoprecipitated and the bound proteins were analyzed by Western immunoblotting using the appropriate antibodies.
Electrophoretic mobility shift assays (EMSAs).
Nuclear extracts prepared from quiescent MCF10A cells infected with Ad variants were incubated with a 5′-end-labeled double-stranded oligonucleotide containing the YY1 binding site of the Myc promoter (5-AGCAAAAGAAAATGGTAGGCGCGCGT-3′; upper strand, −419 to −394 from the P2 cap site) as described previously (30). The reaction mixtures were subjected to 5% nondenaturing polyacrylamide gel electrophoresis at 200 V for 2 h at 4°C, and then the gels were dried and autoradiographed.
Chromatin immunoprecipitaion.
Serum-starved MCF10A cells were treated with 1% formaldehyde at room temperature for 10 to 15 min, and the reaction was stopped by the addition of glycine to a final concentration of 125 mM. Cells were washed twice with ice-cold phosphate-buffered saline, scraped, and suspended in a lysis buffer (50 mM HEPES-KOH [pH 7.9], 10 mM EDTA [pH 8.0], 1% sodium dodecyl sulfate) containing protease inhibitor cocktail (Roche), and then the chromatin was sheared by sonication to an average DNA fragment length of 0.5 to 1 kb. After centrifugation, the soluble cross-linked chromatin was diluted 1:10 in immunoprecipitation buffer (10 mM HEPES-KOH [pH 7.9], 1% Triton X-100, 150 mM NaCl, and protease inhibitors). Chromatin was precleared by incubation with salmon sperm DNA coupled with protein A- or protein G-agarose (50 μl 50% slurry/ml) (Upstate) for 2 h at 4°C. The beads were removed by centrifugation; the precleared chromatin was incubated with antibodies overnight at 4°C, and the DNA-protein complexes were collected using protein A/G agarose. The pellets were washed three times with immunoprecipitation buffer, followed by a wash buffer (10 mM Tris-HCl [pH 8.0], 0.25 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA [pH 8.0]) and Tris-EDTA (pH 8.0). Washed pellets were digested with 50 μg-ml RNase (DNase free; Roche) in Tris-EDTA for 30 min at 37°C. Sodium dodecyl sulfate was added to 0.25% (wt/vol), immunoprecipitated DNA was released by treatment with 250 μg/ml proteinase K for 6 h at 37°C, and formaldehyde cross-links were reversed by heating the samples at 65°C for 6 h. The DNA was extracted with phenol:cholorform:isoamyl alcohol (25:24:1), precipitated with ethanol, and analyzed with primer pairs encompassing the promoter and coding regions of the Myc gene as described in our earlier report (30). The amplified PCR products were resolved by 2% agarose gel electrophoresis and visualized by staining with ethidium bromide. Typically, 4% of the immunoprecipitated sample was used for PCR analysis. The PCR-generated products were between 150 and 300 bp in size. In the case of chromatin histone acetylation experiments, the PCR-generated products were quantified by quantitative real-time PCR using the Applied Biosystems 1200HT system.
RESULTS
E1A interferes in the recruitment of YY1, p300, and HDAC3 to the c-Myc promoter and coding regions in a p300-binding-dependent manner.
In previous studies, we showed that in quiescent cells, transcription factor YY1 binds to the upstream YY1 binding site of the human c-Myc promoter and recruits p300 and HDAC3 to repress c-Myc transcription (30). Recruitment of HDAC3 to the YY1 binding site results in chromatin deacetylation, leading to the repression of c-Myc transcription. The C-terminal half of p300 was found to be sufficient for this activity. Additional studies have demonstrated that YY1, p300, and HDAC3 interact with each other and are present in quiescent cells as a ternary complex. This suggests that the C-terminal region of p300 plays an essential role as an adapter or a corepressor in bridging YY1 and HDAC3. E1A binding to p300 is necessary to induce c-Myc expression in cells arrested by serum starvation. We therefore sought to investigate how E1A affected the binding of the transcription factors to the c-Myc promoter. We showed earlier that WT E1A, but not the E1A mutants RG2 and dl2-36 (see Materials and Methods for the details of mutations), which fail to bind to p300, induces c-Myc transcription (2). These results provided a framework for the studies described below.
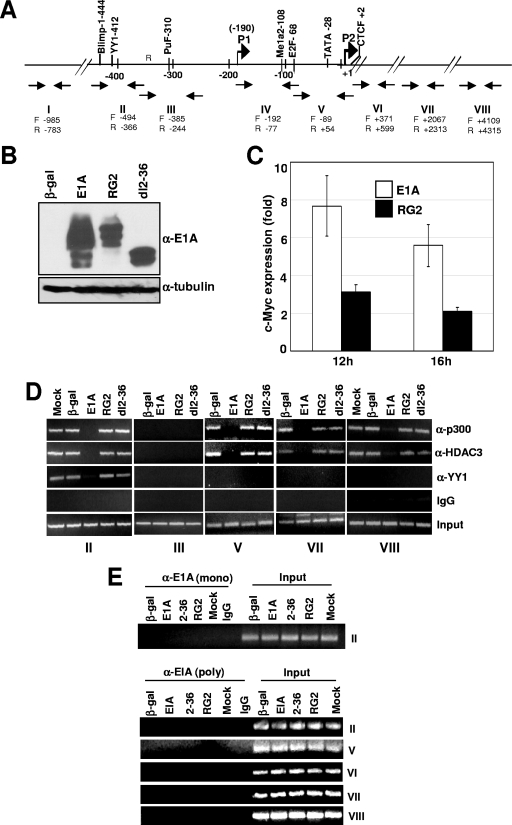
To determine what effect E1A has on the occupancy of the YY1-p300-HDAC3 complex on the YY1 binding site in the Myc promoter, we infected quiescent MCF10A cells (an immortalized nontransformed human breast epithelial cell line) (32) with Ad variants expressing the 243-aa E1A protein (referred to as WT in this article), the E1A RG2 or dl2-36 mutant, or Adb-gal (control). In MCF10A cells, both the RG2 and dl2-36 viruses express the E1A protein at abundant levels at 16 h postinfection (Fig. 1B) (2). Transcription of c-Myc in E1A-expressing cells was confirmed by analyzing Myc RNA using real-time PCR assays as described previously (2). At 16 h after infection, we observed a sixfold increase in c-Myc RNA levels in WT-infected cells over those of cells infected with Adb-gal (control). In contrast, the RG2 mutant showed only a twofold increase (Fig. 1C). At the same time point, cells were harvested and subjected to chromatin immunoprecipitation (ChIP) assays using select primer pairs that encompass the Myc promoter and the coding sequences (Fig. 1A) (30). These primer pairs were chosen based on our previous study which showed that p300 occupies the YY1 binding site and with sequences at the site of the transcriptional initiation and coding regions but not any other upstream promoter regions (30). As shown in Fig. 1D, the presence of p300 at the promoter could be detected at similar levels in mock-infected cells and in cells expressing beta-galactosidase, RG2, and dl2-36. Binding was also observed on the YY1 binding site (region II), the transcriptional initiation region (region V), and the downstream coding regions (regions VII and VIII). Consistent with our previous results, p300 binding was not detected in region III (30). The pattern of HDAC3 occupancy was identical to that of p300, while the YY1 protein could be detected only on the YY1 binding site. In striking contrast, in cells expressing WT E1A, occupancy of all three proteins, YY1, p300, and HDAC3, was abolished on the YY1 binding site. Furthermore, p300 and HDAC3 occupancy was also abolished on the TATA box and the coding regions. Under these experimental conditions, E1A had no effect on the steady-state levels of YY1, p300, and HDAC3 (see Fig. 5). To determine whether E1A occupies these regions of chromatin, we performed ChIP assays using region-specific primer pairs shown in Fig. 1A with a monoclonal or polyclonal antibody directed against E1A. Neither the monoclonal nor the polyclonal antibody coimmunoprecipitated DNA from region II or any other regions shown above, indicating that E1A does not appear to be associated with chromatin at 16 h postinfection (Fig. 1E). The absence of the YY1 protein on the YY1 binding site of the promoter is surprising and suggests either that E1A interferes in YY1 binding to the YY1 binding site or that YY1 binding to the YY1 binding site is dependent on p300 (see Discussion).
FIG. 1.
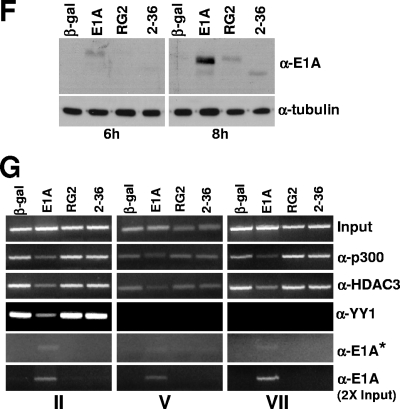
E1A interferes in the recruitment of YY1, p300, and HDAC3 to the Myc promoter and the coding regions. (A) Location of the upstream YY1 binding site on the human c-Myc promoter in relation to the major P2 promoter transcriptional start site and the primer pairs used for ChIP assays. P1 and P2 are the minor and major transcriptional initiation sites, respectively. (B) Expression of the WT and mutant E1A proteins in virus-infected cells. MCF10A cells were serum starved for 32 h and then infected with Ad variants for 16 h, and the cell lysates were subjected to Western immunoblotting using anti-E1A monoclonal antibody (M73; a gift of E. Moran). (C) Analysis of Myc RNA in cells infected with control and E1A viruses. Myc RNA was quantified using a real-time PCR assay as described previously (2). (D) Occupancy of p300, YY1, and HDAC3 on the promoter and coding regions of c-Myc in E1A-expressing quiescent cells. Virus-infected MCF10A cells as above were harvested, and the chromatin was subjected to ChIP assays as described under Materials and Methods. Images of the agarose gels showing the PCR-amplified DNA are shown. The antibodies used are anti-p300 (α-p300) (N15), anti-HDAC3 (α-HDAC3) (B-12), anti-YY1 (α-YY1) (H414), and anti-E1A (α-E1A) (13 S-5 and M73) (all antibodies were from Santa Cruz). The experiment was repeated twice with identical results. (E) Lack of association of E1A on the promoter and coding regions of the Myc chromatin in E1A-expressing cells. Experimental details are as for panel D. Anti-E1A monoclonal antibody M73 (gift of E. Moran) and the anti-E1A polyclonal antibody (Sc-430; Santa Cruz) were used to immunoprecipitate the chromatin. (F) Western blot showing expression of WT and mutant E1A proteins at 6 and 8 h after infection of quiescent MCF10A cells with indicated viruses. Details are as for panel B. (G) Association of E1A with chromatin-bound p300 and dissociation of the p300 complex at 8 h postinfection. Chromatin prepared from virus-infected cells was subjected to ChIP assays with antibodies and primer pairs as shown. The asterisk indicates that the gel image in this panel was obtained with double the exposure time used for other panels. The E1A band in the bottom panel could be detected after taking twice the amount of chromatin used for the other panels.
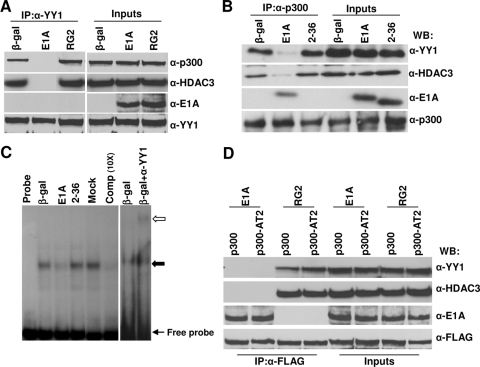
FIG. 5.
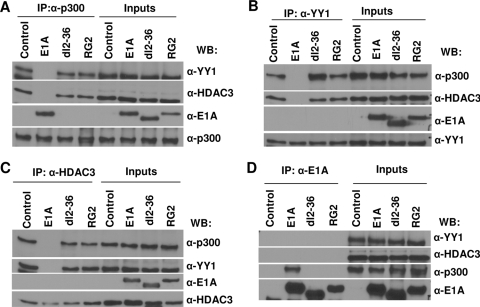
Binding of E1A to p300 interferes in ternary complex formation between p300, YY1, and HDAC3 in vivo, and dissociation of the p300 complex by E1A is not dependent on p300 HAT activity. Serum-starved MCF10A cells were infected with viruses expressing WT E1A or the mutant RG2 or dl2-36 protein for 16 h and harvested, and then the cell lysates were immunoprecipitated using antibodies as shown. The immunoprecipitated (IP) proteins were identified by Western immunoblotting using antibodies as indicated. α-p300, anti-p300; α-HDAC3, anti-HDAC3; α-E1A, anti-E1A; α-YY1, anti-YY1; β-gal, beta-galactosidase. (A) Cell extracts equivalent to 2 mg of protein were immunoprecipitated with anti-YY1 (H414) antibody, followed by Western immunoblotting (WB) with the anti-YY1 (H-10), anti-HDAC3 (B-12), anti-E1A (M73), and anti p300 (RW128) antibodies. For input lanes, 1/20 of the protein used for immunoprecipitation was loaded directly. (B) Immunoprecipitation using anti-p300 (N15) antibody. Other details are the same as for panel A. (C) EMSA showing the effect of E1A on YY1 binding to the YY1 binding site of the Myc promoter. Nuclear extracts were prepared from serum-starved MCF10A cells infected with Ad variants as shown and incubated with the Myc promoter YY1 binding site in vitro. The DNA-YY1 protein complexes (shown by an arrow) were resolved on a 5% nondenaturing polyacrylamide gel as described in Materials and Methods. For the supershift assay, a reaction mixture was incubated in the presence of an anti-YY1 antibody. The open arrow indicates the supershifted band. (D) Cell extracts prepared from cells coinfected with Adp300AT2 (see Materials and Methods) and E1A variants were immunoprecipitated with anti-FLAG antibody (catalog no. F7425; Sigma) and Western immunoblotted using antibodies as indicated. α-FLAG, anti-FLAG antibody.
Since we chose the 16-h time point to look at the effects of E1A on changes in the chromatin-bound p300 complex, it remained possible that E1A could associate with the p300 complex on the chromatin at early times. For example, E1A could bind to the p300 complex as soon as E1A accumulates in the cell and then dissociate the complex. In studies described below, we discovered that E1A initially binds to the ternary complex, stays with the complex briefly, and then dissociates all of the proteins in the complex. Therefore, we examined the effects of E1A on the p300 complex associated with chromatin at early time points. In MCF10A cells, the earliest time point at which we could detect bound E1A was about 8 h. At 6 h, WT E1A was detected at very low levels but mutant proteins were not detectable (Fig. 1F). We also examined the promoter occupancy of YY1, p300, HDAC3, and E1A using chromatin prepared from cells infected for 8 h with primer pairs covering the YY1 binding site, transcription start site, and one of the coding regions, as indicated in Fig. 1G. Data shown in Fig. 1G indicate that at 8 h, as E1A begins to accumulate at low levels, YY1, p300, and HDAC3 occupancy was reduced but not abolished. In the case of E1A, we had to expose the gel for a longer period to detect the E1A band. However, the E1A-related band was clearly detectable when a twofold-increased quantity of chromatin was used for immunoprecipitations. Nonetheless, these results suggest that as E1A accumulates in cells, it binds to chromatin-associated p300, causing the dissociation of YY1, p300, and HDAC3. It is unclear, however, how long it takes to dissociate all of p300, YY1, and HDAC3 from the promoter. Nonetheless, these experiments demonstrate that E1A-induced effects are at the chromatin level. In addition, the chromatin-bound p300 complex on the Myc promoter is very sensitive to E1A binding, which induces the dissociation of all of the proteins present in the complex. In this process, E1A also dissociates from the chromatin.
Dissociation of the YY1-p300-HDAC3 complex from the YY1 site of the c-Myc promoter correlates with chromatin acetylation.
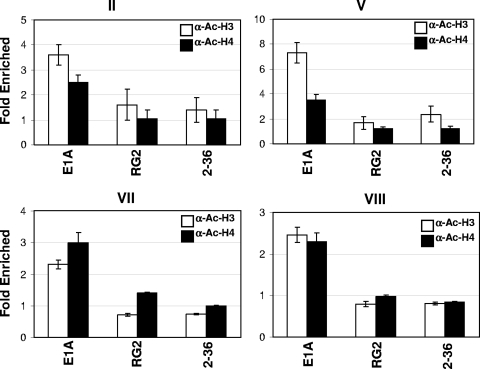
In our previous report, we showed that one consequence of HDAC3 recruitment to the Myc promoter is the deacetylation of chromatin, leading to the transcriptional repression of c-Myc (30). Similarly, in p300-depleted cells, Myc chromatin acetylation is enhanced. Since E1A abolishes the occupancy of HDAC3 on Myc chromatin and transcriptionally activates c-Myc, we sought to determine the acetylation status of chromatin at the Myc promoter. Chromatin prepared from virus-infected cells was immunoprecipitated as described above with anti-acetyl histone H3 and anti-acetyl histone H4 antibodies, and the coimmunoprecipitated DNA was quantified by real-time PCR using primer pairs spanning four different regions of the promoter and the coding regions shown in Fig. 2. As shown in Fig. 2, both anti-acetyl histone H3 and anti-acetyl histone H4 antibodies immunoprecipitated at least twice as much chromatin from WT Ad-infected cells as from Adb-gal-, RG2-, or dl2-36-infected cells. Thus, failure to recruit HDAC3 to the promoter in WT E1A-expressing cells results in increased acetylation of the chromatin. It was reported recently that in E1A-expressing cells, total histone H3 lysine 18 acetylation is decreased by threefold (17). Because the antibodies we used do not recognize H3 lysine 18 acetylation, we do not know at present whether H3 lysine 18 of Myc chromatin is hypoacetylated (see the legend to Fig. 2 for the details of acetylated residues). Together, these results indicate that expression of WT E1A but not that of the p300-binding-defective E1A mutants results in abrogation of YY1, p300, and HDAC3 recruitment to the YY1 binding site. E1A also prevents the recruitment of p300 and HDAC3 to the initiation and coding regions. As a consequence, Myc chromatin is acetylated, resulting in the transcriptional induction of c-Myc.
FIG. 2.
Enhanced acetylation of histones H3 and H4 of Myc chromatin in E1A-expressing cells. Chromatin prepared for ChIP assays as described in Materials and Methods was immunoprecipitated using anti-acetyl H3 (α-Ac-H3) and anti-acetyl H4 (α-Ac-H4) antibodies (catalog no. 06-599 and 06-866, respectively; Upstate). The immunoprecipitated DNA was quantified using real-time PCR with the primer pairs shown in Fig. 1A. The relative increase in real-time PCR values over those for the control samples (cells infected with Adb-gal) is shown. Each value shown is the average for two independent experiments with mean ± standard deviation). The commercial antibodies used in these studies were raised using acetylated peptides. The peptides included amino acids corresponding to 1 to 20 of H3, in which lysines 9 and 14 are acetylated, and 2 to 19 of H4, in which lysine 5, 8, 12, and 16 are acetylated.
E1A binding region in p300 is essential for relief of repression by E1A.
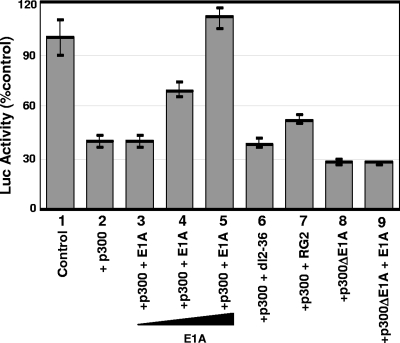
We showed earlier that in virus-infected cells, the WT, but not the E1A variants containing mutations at their N-terminal region, induces c-Myc, implying that binding of E1A to p300 through its N-terminal region is essential for relieving repression (2, 18). To determine whether the interaction of E1A with the CH3 region of p300 is critical for the relief of repression, we performed transient-transfection assays using WT p300 and a p300 mutant in which the E1A binding region is deleted (p300delE1A) (7). Rat12 cells were transfected with a truncated c-Myc promoter-luciferase reporter plasmid containing a p300-responsive minimal promoter fragment (del6) (30), along with plasmids expressing WT p300 or p300delE1A and WT E1A or the E1A mutant RG2 or dl2-36. Transfected cells were maintained in serum-free medium for 36 h and then harvested, and the luciferase activity in the cell lysates was determined. For these studies, the transient assays to measure Myc promoter activity were carried out in quiescent cells to avoid the influence of growth factors on Myc activation. Since transactivation of the Myc promoter is very low in quiescent cells, changes in Myc activities in the presence of different transcriptional effectors were not dramatic. However, they were highly reproducible in multiple independent experiments, and each experiment was carried out in triplicate. Figure 3 shows that in the presence of both WT p300 (bar 2) and p300delE1A (bar 8), Myc reporter activity dropped to less than 40% of that of the control sample. WT E1A reversed the repression caused by WT p300 in a dose-dependent manner (bars 3 to 5), and at the highest concentration of E1A, the reporter activity increased to about 110% of that of the control (compare bar 5 with bar 2). In contrast, the two E1A mutants could not reverse the p300-mediated repression (compare bars 6 and 7 with bar 2), and WT E1A failed to reverse the p300delE1A-mediated repression (compare bar 9 with bar 8). These data suggest that binding of E1A to p300 at the C-terminal region is critical for relieving the repression. Similarly, the N-terminal p300 binding region of E1A is also critical for the relief of repression.
FIG. 3.
Reversal of p300 repression of Myc by E1A is dependent on E1A binding to p300. Rat12 cells were transfected with a Myc promoter-reporter construct containing upstream 640-bp promoter sequences linked to a luciferase promoter-reporter construct (del6), as described in Materials and Methods, along with p300 and E1A expression plasmids as shown. The vector DNA was included wherever appropriate to maintain a constant DNA concentration. After overnight incubation with normal medium, cells were serum starved for 36 h and harvested and the luciferase (Luc) activities quantified. Luciferase units are shown for 5 μg of protein after normalization with Renilla luciferase activity. Samples represented by bars 3, 4, and 5 contained 0.25, 0.5, and 1.0 μg E1A plasmids, respectively. The experiment was repeated at least three times. Each experiment was carried out in triplicate, and the mean values ± standard deviations are shown.
E1A reverses cooperative effects between p300 and HDAC3 or between p300 and YY1 on Myc promoter repression.
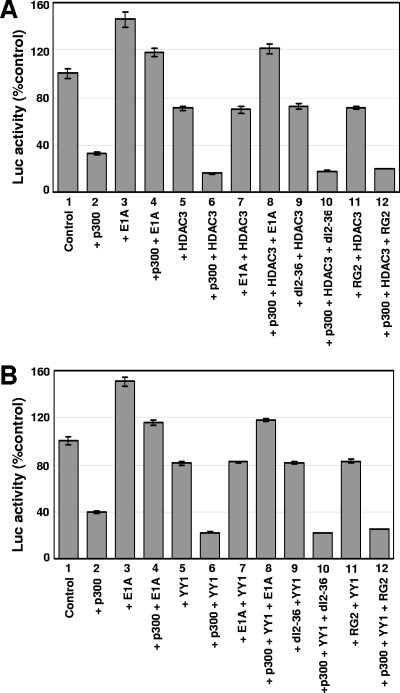
Exogenously added YY1 and HDAC3 cooperate with p300 in repressing the Myc promoter. This is based on the observation that p300 in the presence of YY1 or HDAC3 is more efficient in repressing the Myc promoter in transient assays than p300 alone (30). To determine whether E1A could reverse the repressive effects of exogenously added YY1 and HDAC3 in the presence of p300, transient assays were performed using carefully titrated amounts of p300, HDAC3, and YY1, as described earlier (30). The data shown in Fig. 4A suggest that E1A was able to alleviate the cooperative effect of p300 and HDAC3 on the Myc promoter. For example, the basal activity of the Myc promoter was reduced to about 35% of that of the control in the presence of p300 (bar 2), which was reversed by E1A (compare bar 4 with bar 2). In the presence of HDAC3 alone, Myc repression was moderate (bar 5). However, when p300 and HDAC3 were expressed together, repression of the Myc promoter was more efficient than that with p300 alone (compare bar 6 with bar 2; reporter activity is reduced to about 15% in bar 6). The addition of E1A reversed the repression caused by the combination of p300 and HDAC3 and restored the promoter activity to about 115% (compare bar 8 with bar 6). However, neither dl2-36 nor RG2 was able to reverse the repression caused by the p300 and HDAC3 combined (compare bars 10 and 12 with bar 8).
FIG. 4.
E1A reverses the cooperative effects between p300 and HDAC3 (A) or p300 and YY1 (B) in repressing the Myc promoter in a p300-binding-dependent manner. (A) Reversal of repression of the Myc promoter by E1A in the presence of exogenously added p300 and HDAC3. Rat12 cells were transfected with the del6 promoter construct (0.1 μg) along with p300 and HDAC3 plasmid (1 μg each) and the WT or an E1A mutant (1 μg each). The cells were starved for 36 h, and then the luciferase (Luc) activity was measured. The values are the averages of three independent sets with means ± standard deviations. Conditions for transfection and luciferase assays were as described in Materials and Methods. (B) Reversal of repression of the Myc promoter by E1A in the presence of exogenously added p300 and YY1. Assay conditions are as described for panel A.
E1A also reversed the repression caused by the combined presence of p300 and YY1 in a manner similar to that just described for p300 and HDAC3. For example, Fig. 4B shows that the expression of p300 alone dropped the reporter activity to about 30% of that of the control (compare bar 2 with bar 1). In the presence of YY1 and p300, the promoter activity dropped to about 15% of the control level (compare bar 6 with 2), while YY1 alone did not have a significant effect on reporter activity (bar 5). The cotransfection of WT E1A but not of mutants dl2-36 and RG2 alleviated the repression caused by the combined presence of p300 and YY1 (compare bar 8 with bar 6 for WT E1A and bars 10 and 12 with bar 6 for effects of dl2-36 and RG2). In summary, E1A reverses the inhibition of Myc caused by the cooperative effects of p300 and HDAC3 or p300 and YY1 in a manner that is dependent on binding of E1A to p300.
E1A interferes in protein-protein interaction between YY1, p300, and HDAC3 in vivo.
p300 forms a stable trimolecular complex with HDAC3 and YY1 in vivo and in vitro by a three-way interaction (30). Our failure to detect E1A on the chromatin at 16 h when E1A alleviates p300 repression of c-Myc and also the lack of YY1 occupancy on the YY1 binding site in the presence of E1A suggest that E1A may not be stably associating with promoter-bound YY1 or the promoter-bound YY1-p300-HDAC3 complex to displace the complex from the chromatin. Therefore, we sought to determine whether binding of E1A to p300 interferes in the formation of the trimolecular complex in vivo using coimmunoprecipitation assays. MCF10A cells were made quiescent by serum starvation, infected with Ad viruses for 16 h, and then harvested, and the cell extracts were immunoprecipitated with an anti-p300 antibody. The coimmunoprecipitated proteins were identified by Western immunoblotting. As shown in Fig. 5B, in extracts from cells infected with Adb-gal and dl2-36, the anti-p300 antibody coimmunoprecipitated both YY1 and HDAC3 at similar levels. In contrast, from WT Ad-infected cells, negligible amounts of YY1 and HDAC3 were coimmunoprecipitated with p300. As expected, significant quantities of WT E1A were still coimmunoprecipitated with p300. The results using the RG2 mutant were similar to those shown for dl2-36 (data not shown). A reverse immunoprecipitation experiment using anti-YY1 antibody followed by Western blotting using anti-p300, anti-HDAC3, and anti-E1A confirmed these results (Fig. 5A). These results suggest that binding of E1A to p300 most likely interferes in the formation or the stability of the ternary complex consisting of p300, YY1, and HDAC3 in vivo. In turn, this would account for the finding that YY1 does not occupy the YY1 binding site of the c-Myc promoter in Ad-infected cells.
To test this idea, we used EMSA experiments to determine whether the YY1 protein in cell extracts prepared following virus infection could bind to the YY1 binding site of the Myc promoter in vitro. Nuclear extracts prepared from virus-infected cells were incubated in vitro with a double-stranded oligonucleotide containing the YY1 binding site of the Myc promoter, and the DNA-protein complexes were resolved on nondenaturing polyacrylamide gels using the standard EMSA conditions (see Materials and Methods). As shown in Fig. 5C, a DNA-protein complex was observed in cell extracts prepared from cells infected with Adb-gal or E1A2-36 and mock-infected cells. Addition of an anti-YY1 antibody supershifted the DNA-protein complex, confirming that the complex contained the YY1 protein. In contrast, YY1 present in cell extracts prepared from WT virus-infected cells bound to the YY1 site probe very weakly. We next attempted to determine whether GST-E1A would block binding of the YY1 protein to the YY1 binding site in EMSA. Nuclear extracts prepared from uninfected cells were incubated with the probe in the presence of affinity-purified GST-E1A. The E. coli-based E1A protein did not efficiently interfere with YY1-DNA complex formation in vitro (data not shown). We conclude that in virus-infected cells, E1A prevents the occupancy of the YY1-p300-HDAC3 complex on the YY1 binding site, which is responsible for repression of c-Myc. E1A also interferes in the association of p300 and HDAC3 to the initiation and coding regions of Myc chromatin.
We previously showed that repression of c-Myc by p300 is not related to its intrinsic HAT activity. It was still possible that in E1A-infected cells, the dissociation of the ternary complex by E1A could be dependent on the HAT activity of p300. To determine if this is the case, we repeated the coimmunoprecipitation experiments using an exogenously expressed HAT-defective mutant. In this experiment, we used an Ad vector expressing a FLAG epitope-tagged p300 mutant containing mutations in the HAT domain and thus lacking HAT activity (35). When cell extracts were immunoprecipitated with anti-FLAG antibody, only FLAG-p300-bound proteins should be coimmunoprecipitated. Coimmunoprecipitation with anti-FLAG antibody followed by Western blotting with relevant antibodies showed that E1A bound to both WT and p300AT2 efficiently and dissociated both HDAC3 and YY1 from the complex (Fig. 5D). p300AT2 bound to YY1 and HDAC3 as efficiently as the WT p300. This suggests that dissociation of the complex by E1A is not dependent on p300 HAT activity. However, we cannot rule out the possibility that endogenous p300 HAT activity might have contributed to the dissociation of the complex in trans.
E1A interferes in stable complex formation between p300, YY1, and HDAC3 in vitro.
To gain further insight into the mechanism by which E1A interferes in the protein-protein interactions, we used GST fusion proteins in in vitro assays to investigate whether or not E1A would interfere in the formation of a complex containing p300, YY1, and HDAC3. The three GST fusion proteins were mixed and incubated together on ice for 2 h in the presence of WT E1A or one of the two E1A mutants that do not bind to p300 (all are GST fusion proteins). Following incubation, the samples were diluted in the lysis buffer as described in Materials and Methods and immunoprecipitated with the antibodies as appropriate, and the coimmunoprecipitated proteins were identified by Western immunoblotting. For p300, we used the C-terminal aa 1572- to-2370 region of p300 since our previous studies showed this portion of p300 is sufficient for the interaction of YY1 and HDAC3 (30). This fragment of p300 also contains the E1A binding region (7). GST was used as a control in all in vitro binding assays. As shown in Fig. 6A, immunoprecipitations using anti-p300 antibodies showed that the C-terminal region of p300 formed a complex with both YY1 and HDAC3 in the control incubations, indicating the formation of a stable trimolecular complex in vitro (control lane), a result that is consistent with our previous results (30). Anti-p300 antibody also brought down YY1 and HDAC3 in the presence of RG2 and dl2-36 but not in the presence of WT E1A. When WT E1A was present in the incubation mixture, anti-p300 antibody brought down only the E1A protein. These results suggest that E1A binding to p300 through its N-terminal sequences interferes with binding of YY1 and HDAC3 to p300. These results were further confirmed using antibodies directed against YY1 and HDAC3 to immunoprecipitate complexes formed in vitro. YY1- and HDAC3-specific antibodies were able to coimmunoprecipitate complexes containing p300 and HDAC3 and p300 and YY1, respectively, from the control sample and the samples containing the two E1A mutants (Fig. 6B and C). When WT E1A was present in the incubation mixture, HDAC3 did not coimmunoprecipitate with antibodies to YY1 (Fig. 6B) and YY1 did not coimmunoprecipitate with antibodies to HDAC3 (Fig. 6C), since these proteins were dissociated from the complex by 2 h. The E1A antibody could coimmunoprecipitate p300 only from cells expressing WT E1A (Fig. 6D) but not from cells expressing the two E1A mutants. Together, these results indicate that E1A prevents the formation of stable complexes containing p300, YY1, and HDAC3.
FIG. 6.
E1A interferes in complex formation between p300, YY1, and HDAC3 in vitro. GST fusion proteins containing YY1, HDAC3, and the C-terminal region of p300 (0.5 μg each) and E1A (1.0 μg) were mixed, incubated in vitro on ice for 2 h, and immunoprecipitated, followed by Western immunoblotting using antibodies as shown. Sources of antibodies are as indicated in legends to Fig. 1D and 5A; antibody abbreviations are as in the legend to Fig. 5. For input lanes, proteins at 1/20 of the amounts used for the assays were loaded directly.
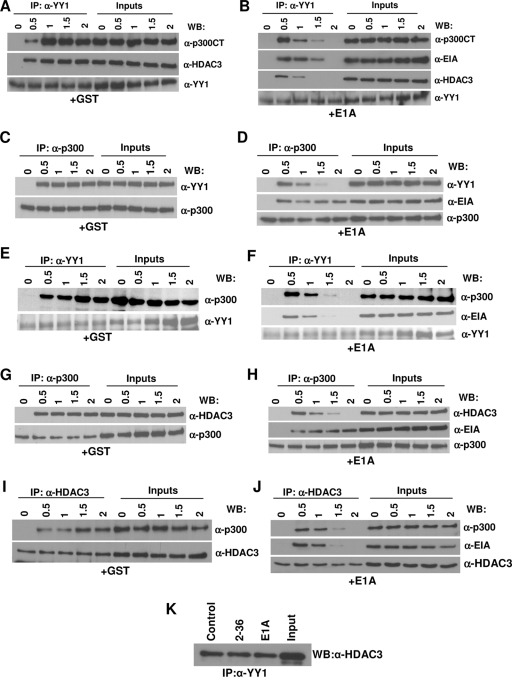
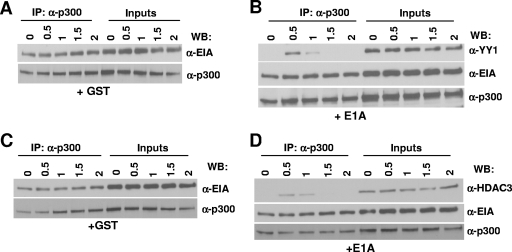
Since the effect of E1A on the ternary complex was determined after 2 h of incubation in the above experiment, it was possible that E1A might transiently associate with the trimolecular complex, resulting in subsequent dissociation of the other proteins. In this case, one should be able detect a complex containing E1A and the other three proteins early in a time course experiment. To address this possibility, an experiment was carried out in which GST derivatives of p300, HDAC3, YY1, and E1A were incubated together on ice for various lengths of time before immunoprecipitation. When anti-YY1 antibody was used, control experiments showed that the three proteins formed a complex by 30 min of incubation and that the complex was stable even after 2 h (Fig. 7A). In contrast, in the presence of GST-E1A, a significant amount of E1A was associated with the p300-YY1-HDAC3 complex but only at the 30-min and 1-h time points (Fig. 7B). The complex began to dissociate by 1 h, and none of the associated proteins could be detected at the 2-h time point.
FIG. 7.
Binding of E1A to p300 in the p300 protein complexes leads to the dissociation of YY1 and HDAC3 from the complex in vitro. (A and B) Association of E1A with the trimolecular complex at early time points during incubation and dissociation of the complex as a function of time. p300, YY1, HDAC3, and E1A (B) or GST (A) were mixed and incubated on ice. At the indicated time points, proteins were immunoprecipitated with an anti-YY1 antibody, and the coimmunoprecipitated proteins were identified by Western blotting with antibodies as indicated. α-p300CT, anti-p300CT antibody; α-HDAC3, anti-HDAC3 antibody; α-YY1, anti-YY1 antibody. (C to F) Effect of E1A on YY1-p300 complex formation. YY1, p300, and GST (C and E) or E1A (D and F) were mixed, incubated on ice, and coimmunoprecipitated with anti-p300 (C and D) or anti-YY1 (E and F) antibodies at the indicated time points, followed by Western immunoblotting with antibodies as shown. (G to J) Effect of E1A on HDAC3-p300 complex formation. HDAC3, p300, and GST (G and I) or E1A (H and J) were incubated on ice and coimmunoprecipitated with antibodies, followed by Western immunoblotting with antibodies as shown. (K) HDAC3-YY1 complex formation is not affected by E1A. GST fusion proteins containing YY1 and HDAC3 (0.5 μg each) were mixed with GST (control), WT E1A, or dl2-36 (1 μg each), incubated for 2 h on ice, and then immunoprecipitated with anti-YY1 antibody and Western immunoblotted with anti-HDAC3 antibody.
To further define the effect of E1A on these protein-protein interactions, we used incubations with only two of the cellular proteins at a time. The incubations included YY1 and p300 (Fig. 7C to F), HDAC3 and p300 (Fig. 7G to J), and HDAC3 and YY1 (Fig. 7K) in the presence and absence of E1A. All immunoprecipitations with the exception of results shown in Fig. 7K were carried out in a reciprocal manner using antibodies specific for either protein. In the absence of E1A, p300 and YY1 formed a stable complex by 30 min which remained intact even after 2 h (Fig. 7C and E), whereas in the presence of E1A, the association of p300 and YY1 could be detected at only the 30-min and 1-h time points (Fig. 7D and F). E1A bound to the YY1-p300 complex by 30 min (Fig. 7D and F) and remained associated with p300 even after 2 h (Fig. 7D). YY1 began to dissociate from the complex by 1 h, and by 2 h, the YY1-p300 complex could not be detected (Fig. 7D and F) and YY1 was not bound to E1A (Fig. 7F). Similarly, by 30 min of incubation, a complex containing p300-HDAC3-E1A could be detected (Fig. 7H and J). HDAC3 began to dissociate from the complex by 1 h, and by 2 h of incubation, HDAC3 dissociation was complete whereas E1A continued to be bound to p300 (Fig. 7H and J).
To determine whether the YY1-HDAC3 complex would be affected by the presence of E1A, we incubated YY1 and HDAC3 in the presence of GST or E1A variants in vitro for 2 h, and then the complex formation was tested as described above. Data presented in Fig. 7K suggest that YY1-HDAC3 complex formation was not affected by E1A. Because the binding of YY1 to E1A is very weak (21), we observed the YY1-E1A interaction only when high concentrations of GST-E1A were used (5 μg; data not shown). Even at these high concentrations, the YY1-HDAC3 association was still observed. In contrast, an E1A-HDAC3 interaction could not be detected even at high concentrations of E1A (data not shown). Thus, binding of E1A to p300 is primarily responsible for the disruption of the trimolecular complex.
We also determined whether the preformed p300-E1A complex would prevent other proteins from binding to the complex. p300 and E1A were mixed and incubated for 2 h on ice before the addition of YY1 and HDAC3. Results shown in Fig. 8A to D suggest that both YY1 and HDAC3 bind to the p300-E1A complex initially, although binding appears to be somewhat weaker than that observed when all of the proteins were mixed simultaneously (results shown in Fig. 7). E1A continued to be bound to p300 even after 2 h of incubation (a total of 4 h of incubation), while YY1 and HDAC3 bound to the p300-E1A complex briefly and then dissociated from the complex (Fig. 8B and D).
FIG. 8.
Preformed p300-E1A complex still allows transient association of YY1 or HDAC3 with the complex. (A to D) p300 and E1A were preincubated on ice for 2 h to form a complex, and then either GST (A and C) or YY1 (B) and HDAC3 (D) were added. At the indicated time points, the proteins were immunoprecipitated (IP) using anti-p300 antibodies and Western immunoblotted (WB) with antibodies as shown. For antibody abbreviations, see the legend to Fig. 5.
In summary, the in vitro experiments using GST fusion proteins suggest that E1A can bind p300 at the level of the ternary complex and then become stably associated with p300, eventually resulting in the dissociation of both YY1 and HDAC3 from the complex.
DISCUSSION
In a previous article, we showed that p300 represses c-Myc expression through the upstream YY1 binding site of the c-Myc promoter. p300 provides a corepressor function in that the C-terminal half of p300 functions as an adaptor to bridge YY1 and HDAC3 (30). In other studies, we have shown that the 243-aa transforming E1A protein induces c-Myc in quiescent cells and that the E1A-induced DNA synthesis in quiescent cells is the result of c-Myc induction (2). E1A mutants with mutations in their N-terminal region do not bind to p300, and these mutants are also impaired in their capacity to induce DNA synthesis and cell transformation (2). Activation of c-Myc is critical for both DNA synthesis and cell transformation. Thus, our previous studies provided a molecular explanation for the failure of E1A mutants that do not bind p300 to induce S phase and transform cells. The present studies were undertaken with the aim of understanding the molecular mechanism by which E1A induces c-Myc in quiescent cells.
In this article, we have shown that when the E1A protein is expressed in quiescent human cells, significant levels can be detected by about 8 h. As E1A begins to accumulate, it associates with chromatin-bound p300, and at the same time, YY1, p300, and HDAC3 begin to dissociate from the YY1 binding site. p300 and HDAC3 also begin to dissociate from the transcription initiation and coding regions. However, at 16 h after infection, when E1A accumulates to abundant levels, all three proteins of the ternary complex are dissociated from the promoter, and in the process, E1A also dissociates from the complex. Occupancy of the p300 complex on the YY1 binding site and other regions of the Myc chromatin is normal in cells expressing two E1A mutants that do not bind p300, suggesting that binding of E1A to p300 through its N-terminal region prevents recruitment of these proteins to the promoter. Transient-transfection studies using expression constructs also support this observation. For example, E1A reverses the repression of c-Myc mediated by WT p300 but not by a p300 mutant in which the E1A binding site is deleted. Similarly, E1A mutants that do not bind to p300 are unable to reverse the repression of c-Myc by p300. Earlier we demonstrated that p300, HDAC3, and YY1 are present as a ternary complex in vivo and that the complex is formed by a three-way interaction (30). In the present article, we have shown that in virus-infected cells, WT E1A interferes in the formation of the ternary complex. E1A is the only protein that associated stably with p300 in E1A-expressing cells (Fig. 5). We also ruled out the possibility that in E1A-infected cells, p300 HAT activity plays a role in the E1A-mediated dissociation of the complex, because the properties of a HAT-defective p300 mutant were identical to those of WT p300 in the dissociation of the complex.
The changes we observed in p300, YY1, and HDAC3 occupancy on the Myc promoter were not due to forced entry of E1A-expressing cells into S phase, since these changes were also detected in the presence of a Myc inhibitor which blocks Myc DNA binding activity (34a) and hence S-phase entry of E1A-expressing cells (data not shown). This Myc inhibitor does not inhibit Myc transcription significantly (34a).
These results did not address whether binding of E1A to p300 prevents the interaction of YY1 and HDAC3 with p300. In order to gain further insight into the effect of E1A on the formation of the p300-YY1-HDAC3 complex, we employed in vitro assays using GST fusion proteins in various combinations. Interestingly, a time course study revealed that E1A binds to the trimolecular complex initially since E1A binding to the complex can be observed in 30- and 60-min incubations (Fig. 7). At later times, the only protein that remains bound to p300 is E1A. This is also true when two-protein interactions (p300 and YY1 or p300 and HDAC3) are tested in the presence of E1A (Fig. 7). In each case, E1A continues to be associated with p300 while HDAC3 or YY1 is dissociated from the complex by 2 h. These results agree with the ChIP results shown in Fig. 1G. At an early time point when E1A can be detected in virus-infected cells, E1A bound to chromatin-associated p300. Along with the association of E1A on chromatin, we also observed reduced occupancy of YY1, p300, and HDAC3 on the YY1 binding site and also reduced association of p300 and HDAC3 on other regions of Myc chromatin. A previous study showed only weak binding of E1A to YY1 (21), and we have confirmed this result. At the concentration of E1A used for binding assays (1 μg), we could not detect significant interaction of E1A with YY1. We were also unable to detect E1A binding to HDAC3 at this E1A concentration. At a fivefold-higher concentration, we could detect an interaction between YY1 and E1A but not between HDAC3 and E1A. Since all of our in vitro binding assays were carried out with 1 μg of E1A, YY1-E1A interactions may not be relevant for our observations. Thus, the association of E1A with the ternary complex must occur through the E1A binding site on p300. Binding of E1A to p300 could result in the displacement of YY1 and HDAC3 from the complex. Importantly, it appears that E1A can dissociate other proteins from the p300 complex both at the chromatin level and in vitro.
The observation that E1A can associate with the trimolecular complex at the chromatin level and in the time course experiment (Fig. 7) in vitro suggests that the E1A binding region of p300 in the complex is available for E1A to interact with p300. This is consistent with the observation that the p300 mutant in which the E1A binding region is deleted is still active in the repression assays (Fig. 3). Currently we do not know what molecular alterations take place in p300 when E1A binds to p300 such that YY1 and HDAC3 cannot associate with the complex after some time. It is conceivable that binding of E1A to the p300-YY1-HDAC3 complex through the E1A binding region of p300 may destabilizes the complex such that other proteins can no longer associate with p300.
The absence of YY1 on the YY1 binding site in E1A-expressing cells shown here is noteworthy. We previously showed that in p300-depleted cells, occupancy of YY1 on the YY1 binding site is much reduced. Thus, YY1 occupancy on the YY1 binding site, at least in the case of the c-Myc promoter, may be dependent on association of YY1 with p300. Others have suggested that YY1 may be present in cells as a complex with p300 (20). Thus, in E1A-expressing cells, when interaction of YY1 with p300 is disrupted, YY1 is unable to bind stably to the YY1 binding site. Similarly, a lack of p300 on the transcription initiation and coding regions may also be related to E1A binding to p300 and disrupting the interaction of p300 with chromatin factors (14). However, we note that this may not be a common property of E1A. There are examples in which transcription factors bind to cognate sites in E1A-expressing cells. Others have shown that transcription factors are capable of binding their cognate sites in vivo. For example, E2F1, -2, and -3 bind to promoters of cell cycle-related genes in E1A-expressing cells (11). Similarly, the corepressor CtBP2 binds to E-cad and c-Fos promoters in E1A-expressing cells (36). In E1A-expressing cells, general transcription factors, such as TBP and RNA polymerase II, must bind to DNA to activate transcription.
The mechanism by which E1A relieves repression of c-Myc by p300 resembles that of the relief of Rb-mediated repression of E2F by E1A. In quiescent cells, E2F is kept repressed by the chromatin complexes containing Rb family proteins and HDAC1. E1A binds to the Rb family proteins through its pocket domain, dissociates the complex, and releases E2F (23). ChIP analysis of the cell cycle-related promoters has shown that E1A associates with p130 and replaces E2F4 with other E2F family members (11). In a similar manner, the small E1A protein binds to p300 through its CH3 domain and dissociates the YY1 from the promoter. In both cases, E1A binds to coregulators at the level of chromatin.
Another important question is whether c-Myc induction is essential for other virus-encoded oncogenes to induce S phase. Simian virus 40 large T antigen is known to induce S phase in quiescent cells, and similarities have been reported in the mechanisms by which SV40 large T and E1A induce S phase (16). SV40 large T also interacts with p300, although this interaction is indirect in that large T and p300 are bridged by p53 (4). We recently showed that the indirect interaction of large T with p300 results in the induction of c-Myc (31). Thus, it will be interesting to determine whether large-T-mediated induction of c-Myc involves the YY1 transcription factor and whether large T has a capacity to dissociate the p300-YY1-HDAC3 ternary complex discussed here.
Recent studies suggest that c-Myc targets a large number of cellular genes to activate transcription (5). c-Myc plays a pivotal role in a number of pathways, including those involved in differentiation, cell proliferation, and development (6). Since deregulation of c-Myc is associated with a number of human cancers, stringent regulation of c-Myc in normal cells is critical, and the chromatin repressor complexes may play an important role in this regulation. Thus, in addition to the p300-containing complexes described by us, there may be other chromatin complexes present in the cell to negatively regulate c-Myc. For example, a recent study reported negative regulation of c-Myc by distinct SWI/SNF chromatin remodeling complexes during differentiation-associated cell cycle arrest (28). It remains to be determined whether E1A would use the mechanism similar to that described here to relieve the repression of c-Myc by the complexes described above.
A recent analysis of the effects of E1A on cellular gene expression on a global scale showed that in E1A-expressing cells, at late times, p300 remains associated with the promoters of the proliferation-related genes to acetylate chromatin (8, 17). In the case of c-Myc, we showed that at 16 h postinfection, p300 is dissociated from the chromatin. Perhaps other proteins in the cell acetylate chromatin at the Myc promoter in the absence of p300. Since their studies also showed a threefold reduction of total of histone H3 lysine 18 acetylation (17), we do not know whether H3 lysine 18 of Myc chromatin is hypoacetylated at present. Our results of c-Myc regulation in E1A-expressing cells are consistent with our previous observations about c-Myc upregulation in p300 knockdown cells. In p300-depleted cells, YY1 and HDAC3 are dissociated from the promoter when c-Myc is upregulated.
At present we do not know whether there are other cellular genes involved in the induction of S phase that are repressed by p300 in quiescent cells and whether E1A uses the mechanism described here to induce those genes. These studies are in progress. c-Fos is one of the early genes whose activation is critical for the induction of S phase in quiescent cells, and studies have shown that its promoter contains functional YY1 binding sites (26). However, we showed earlier that p300 overexpression leads to repression of c-Myc but not c-Fos, indicating that the role of p300 in YY1-mediated repression of gene expression may not be universal (1).
Acknowledgments
We are particularly grateful to Timothy Foster of LSUHSC, New Orleans, for allowing the first author of this article to perform some of the immunoprecipitation studies at his current location after the first submission of the manuscript. We are also grateful to E. Seto, D. Livingston, and Y. Shi for providing us with the plasmids, E. Moran for providing us with the viruses and E1A antibodies, and K. Rundell and L. Laiminis for critical reading of the manuscript.
This work was supported by Public Health Service grant CA74403 from the National Cancer Institute.
Footnotes
Published ahead of print on 11 March 2009.
REFERENCES
- 1.Baluchamy, S., H. N. Rajabi, R. Thimmapaya, A. Navaraj, and B. Thimmapaya. 2003. Repression of c-Myc and inhibition of G1 exit in cells conditionally over expressing p300 that is not dependent on its histone acetyltransferase activity. Proc. Natl. Acad. Sci. USA 1009524-9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baluchamy, S., N. Sankar, A. Navaraj, E. Moran, and B. Thimmapaya. 2007. Relationship between E1A binding to cellular proteins, c-myc activation and S-phase induction. Oncogene 26781-787. [DOI] [PubMed] [Google Scholar]
- 3.Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 247673-7685. [DOI] [PubMed] [Google Scholar]
- 4.Borger, D. R., and J. A. DeCaprio. 2006. Targeting of p300/CREB binding protein coactivators by simian virus 40 is mediated through p53. J. Virol. 804292-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang, C. V., L. M. Resar, E. Emison, S. Kim, Q. Li, J. E. Prescott, D. Wonsey, and K. Zeller. 1999. Function of the c-Myc oncogenic transcription factor. Exp. Cell Res. 25363-77. [DOI] [PubMed] [Google Scholar]
- 7.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8869-884. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari, R., M. Pellegrini, G. A. Horwitz, W. Xie, A. J. Berk, and S. K. Kurdistani. 2008. Epigenetic reprogramming by adenovirus e1a. Science 3211086-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frisch, S. M., and J. S. Mymryk. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 3441-452. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 target is an essential E1A transformation target. Cell 106297-307. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh, M. K., and M. L. Harter. 2003. A viral mechanism for remodeling chromatin structure in G0 cells. Mol. Cell 12255-260. [DOI] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 141553-1577. [PubMed] [Google Scholar]
- 14.Guermah, M., V. B. Palhan, A. J. Tackett, B. T. Chait, and R. G. Roeder. 2006. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125275-286. [DOI] [PubMed] [Google Scholar]
- 15.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 2811509-1512. [DOI] [PubMed] [Google Scholar]
- 16.Helt, A. M., and D. A. Galloway. 2003. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 24159-169. [DOI] [PubMed] [Google Scholar]
- 17.Horwitz, G. A., K. Zhang, M. A. McBrian, M. Grunstein, S. K. Kurdistani, and A. J. Berk. 2008. Adenovirus small e1a alters global patterns of histone modification. Science 3211084-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolli, S., A. M. Buchmann, J. Williams, S. Weitzman, and B. Thimmapaya. 2001. Antisense-mediated depletion of p300 in human cells leads to premature G1 exit and up-regulation of c-MYC. Proc. Natl. Acad. Sci. USA 984646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 198123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J. S., K. M. Galvin, R. H. See, R. Eckner, D. Livingston, E. Moran, and Y. Shi. 1995. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 91188-1198. [DOI] [PubMed] [Google Scholar]
- 21.Lewis, B. A., G. Tullis, E. Seto, N. Horikoshi, R. Weinmann, and T. Shenk. 1995. Adenovirus E1A proteins interact with the cellular YY1 transcription factor. J. Virol. 691628-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litovchick, L., S. Sadasivam, L. Florens, X. Zhu, S. K. Swanson, S. Velmurugan, R. Chen, M. P. Washburn, X. S. Liu, and J. A. DeCaprio. 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26539-551. [DOI] [PubMed] [Google Scholar]
- 23.Macaluso, M., M. Montanari, and A. Giordano. 2006. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene 255263-5267. [DOI] [PubMed] [Google Scholar]
- 24.Montell, C., E. F. Fisher, M. H. Caruthers, and A. J. Berk. 1982. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature 295380-384. [DOI] [PubMed] [Google Scholar]
- 25.Moran, E. 1994. Cell growth control mechanisms reflected through protein interactions with the adenovirus E1A gene products. Semin. Virol. 5327-340. [Google Scholar]
- 26.Murphy, D. J., S. Hardy, and D. A. Engel. 1999. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol. 192724-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mymryk, J. S., and S. T. Bayley. 1993. Multiple pathways for gene activation in rodent cells by the smaller adenovirus 5 E1A protein and their relevance to growth and transformation. J. Gen. Virol. 742131-2141. [DOI] [PubMed] [Google Scholar]
- 28.Nagl, N. G., Jr., D. R. Zweitzig, B. Thimmapaya, G. R. Beck, Jr., and E. Moran. 2006. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 661289-1293. [DOI] [PubMed] [Google Scholar]
- 29.Rajabi, H. N., S. Baluchamy, S. Kolli, A. Nag, R. Srinivas, P. Raychaudhuri, and B. Thimmapaya. 2005. Effects of depletion of CREB-binding protein on c-Myc regulation and cell cycle G1-S transition. J. Biol. Chem. 280361-374. [DOI] [PubMed] [Google Scholar]
- 30.Sankar, N., S. Baluchamy, R. K. Kadeppagari, G. Singhal, S. Weitzman, and B. Thimmapaya. 2008. p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene 275717-5728. [DOI] [PubMed] [Google Scholar]
- 31.Singhal, G., R. K. Kadeppagari, N. Sankar, and B. Thimmapaya. 2008. Simian virus 40 large T overcomes p300 repression of c-Myc. Virology 377227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soule, H. D., T. N. Maloney, S. R. Wolman, W. D. J. Peterson, R. Brenz, C. M. McGrath, J. Russo, R. J. Pauley, R. F. Jones, and S. C. Brooks. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line MCF-10. Cancer Res. 506075-6086. [PubMed] [Google Scholar]
- 33.Stein, R. W., M. Corrigan, P. Yaciuk, J. Whelan, and E. Moran. 1990. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J. Virol. 644421-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, H. G., Y. Rikitake, M. C. Carter, P. Yaciuk, S. E. Abraham, B. Zerler, and E. Moran. 1993. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol. 67476-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Yin, X., C. Giap, J. S. Lazo, and E. V. Prochownik. 2003. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 226151-6159. [DOI] [PubMed] [Google Scholar]
- 35.Zeng, S. X., Y. Jin, D. Kuninger, P. Rotwein, and H. Lu. 2003. The acetylase activity of p300 is dispensable for MDM2 stabilization. J. Biol. Chem. 2787453-7458. [DOI] [PubMed] [Google Scholar]
- 36.Zhao, L. J., T. Subramanian, S. Vijayalingam, and G. Chinnadurai. 2007. PLDLS-dependent interaction of E1A with CtBP: regulation of CtBP nuclear localization and transcriptional functions. Oncogene 267544-7551. [DOI] [PMC free article] [PubMed] [Google Scholar]