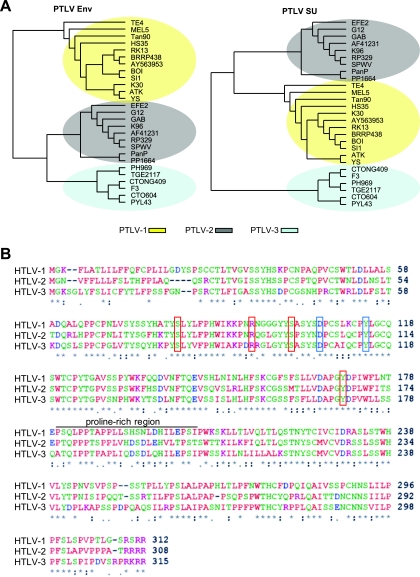
FIG. 1.
HTLV-3 Env and HTLV-3 SU belong to different phylogenetic clades than the HTLV-1 and HTLV-2 Env and SU but display a similar domain organization. (A) Phylogenetic trees resulting from the analysis of 27 sequences of either the entire env gene (1,479 nt) (left) or the portion that encodes the SU (951 nt) (right). The sequences were aligned using ClustalW (in DAMBE program). The trees shown were generated in PAUP using the maximum likelihood method. (B) The SU amino acid consensus sequences of HTLV-1, HTLV-2, and HTLV-3 were aligned using ClustalW. An asterisk indicates identical residues, a colon indicates conserved substitutions, and a period indicates semiconserved substitutions. Red boxes, residue involved in Env-mediated fusion and infection (S81, R94, S101, Y170); blue boxes, residues involved in GLUT-1 interactions (D106 and Y114).