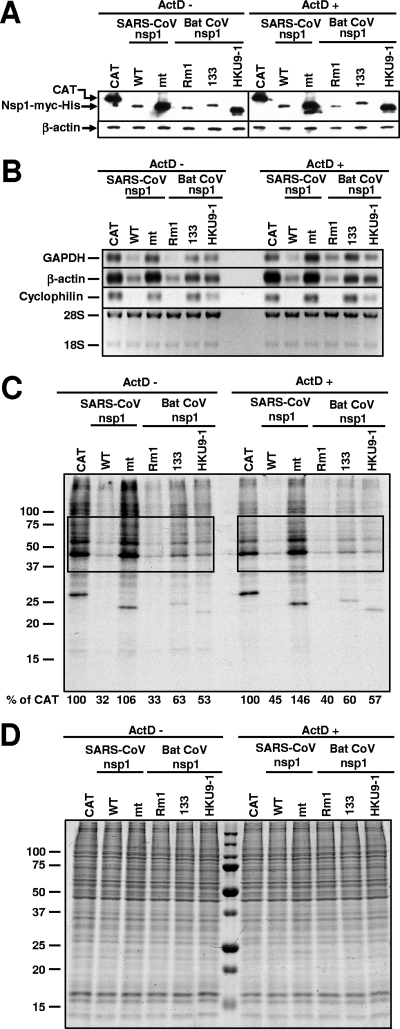
FIG. 3.
Effect of bat CoV nsp1 expression on host mRNA stability and protein synthesis. 293 cells were independently transfected with in vitro-synthesized RNA transcripts encoding CAT (CAT), SARS-CoV nsp1 (WT), SCoVnsp1-mt (mt), Rm1 nsp1 (Rm1), 133 nsp1 (133), or HKU9-1 nsp1 (HKU9-1) protein. One hour after RNA transfection, cells were incubated in the absence of ActD (ActD −) or the presence of ActD (ActD +). (A) At 8 h posttransfection, total proteins were extracted. Western blot analysis was performed using anti-myc or anti-β-actin antibodies. (B) Total RNAs were extracted at 8 h posttransfection and used for Northern blot analysis with riboprobes specific for GAPDH, β-actin, and cyclophilin. The abundance of 28S and 18S rRNAs is shown at the bottom. (C and D) Cells were radiolabeled with 20 μCi/ml of [35S]methionine from 6.5 to 7 h after the ActD addition. Equivalent amounts of intracellular proteins were analyzed by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis. (C) The gel was exposed to X-ray film. Phosphorimager analysis was performed to determine the level of host protein synthesis, and the numbers below the lanes represent the percentages of radioactivity relative to that for cells transfected with CAT RNA (% of CAT). The boxes indicate the regions of the gel used for phosphorimager analysis. Representative data from two independent experiments are shown. (D) The gel was stained with colloid Coomassie blue.