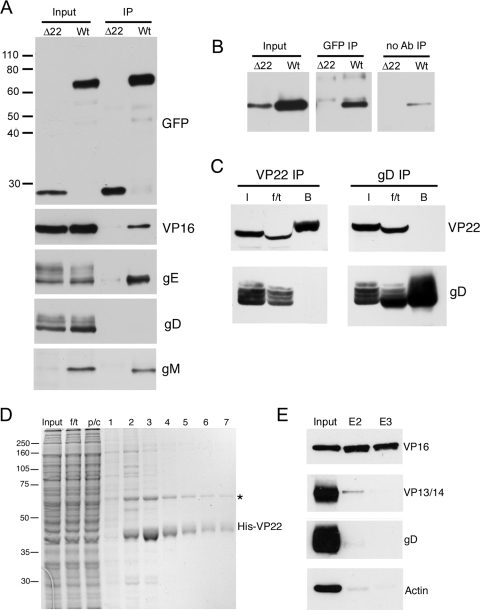
FIG. 1.
Binding partners of VP22 in infected cells. (A) RIPA extracts were made from Vero cells infected with HSV-1 expressing GFP-VP22 (Wt) or HSV-1 expressing GFP in place of VP22 (Δ22), and immunoprecipitations were carried out with a polyclonal anti-GFP antibody. The resulting immunocomplexes (IP) were analyzed by SDS-PAGE, followed by Western blotting with antibodies for GFP, VP16, gE, gD, and gM. (B) Immunoprecipitations were carried out as for panel A, but samples in loading buffer were heated to 56°C rather than boiled before analysis by SDS-PAGE, followed by Western blotting for gM. A control immunoprecipitation in the absence of antibody was also carried out (no Ab). (C) Vero cells infected with strain 17 of HSV-1 were subjected to immunoprecipitation for VP22 and gD by the method of Chi and coworkers (4). I, input; f/t, flowthrough; B, bound. NB: the presence of the f/t sample on these gels has distorted the appearance of some bands. (D) Polyhistidine-tagged VP22 purified from baculovirus-infected Sf9 cells was bound to Ni-NTA beads and incubated for 1 h with a precleared cell extract from 108 strain 17-infected Vero cells. The beads were washed twice, and then bound material was eluted in 500-μl aliquots. Samples were analyzed by SDS-PAGE, followed by Coomassie blue staining. f/t, flowthrough; p/c, precleared extract. The asterisk denotes the major 65-kDa VP22 binding protein.(E) Samples of the input extract and eluates 2 and 3 (E2 and E3) were analyzed by Western blotting with antibodies for VP16, VP13/14, gD, and actin. The values on the left are molecular sizes in kilodaltons.