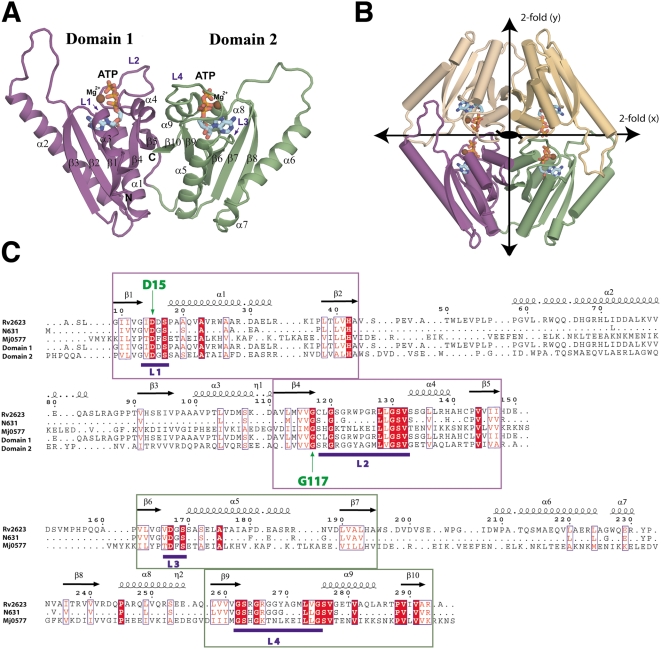
Figure 6. Structure and phylogeny of Rv2623 from M. tuberculosis.
(A,B) A ribbon representation of the Rv2623 monomer (A) and dimer (B) with bound ATP (sticks) and Mg2+ (chocolate spheres). The three, mutually perpendicular pseudo-two-fold axes of the dimer are represented by lines with double arrows (along x, y) and a central ellipse (along z). The atoms of the bound ATP are colored cyan (carbon), red (oxygen), blue (nitrogen), and orange (phosphorus) in (A) and (B). (C) A structure-based sequence alignment of Rv2623, the N631 subfamily consensus, Methanococcus jannaschii protein 0577 (MJ0577), and domains 1 and 2 of Rv2623. Invariant residues in the alignment (>85% conserved in N631) are shaded in bold red and similarities are boxed in blue but left unshaded. Regions with consensus ATP binding motifs comprising L1/L2 (domain 1) and L3/L4 (domain 2) are colored dark violet and smudge, respectively. The positions of the mutated amino acids (D15, G117) are indicated in green. The structure-based sequence alignment was produced using ESPript and the structural representations were produced using PyMOL.