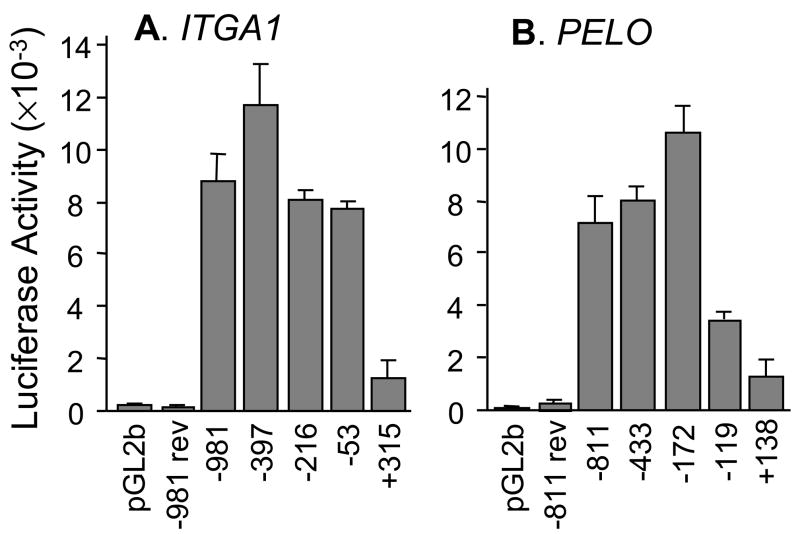
FIGURE 4.
Deletion analysis of the proximal 5′-regulatory region of A) ITGA1 or B) PELO.
A) A series of reporter plasmids containing fragments of the 5′-region of human ITGA1 extending from −981, −397, −216, −53, or +315 and through +455 were inserted upstream from the LUC reporter gene in pGL2basic (pGL2b) and transfected into Dami cells. Baseline activity was measured with the pGL2basic (pGL2b) vector alone. Co-transfection of the PRL-TK plasmid was used to normalize for transfection efficiency. Each vertical bar represents the mean +/− 1 SD for three independent experiments. For each construct depicted on the abscissa, relative luciferase activity is indicated on the ordinate. −981 rev represents the entire 5′-sequence from −981 to +455 inserted in the reverse orientation.
B) A series of reporter plasmids containing fragments of the 5′-region of human PELO extending from −811, −433, −172, −119, or +138 and through +180 were inserted upstream from the LUC reporter gene in pGL2basic (pGL2b) and transfected into Dami cells. Baseline activity was measured with the pGL2basic (pGL2b) vector alone. Each vertical bar represents the mean +/− 1 SD for four independent experiments. For each construct depicted on the abscissa, relative luciferase activity is indicated on the ordinate. −811 rev represents the entire 5′-sequence (−811/+180) inserted in the reverse orientation.