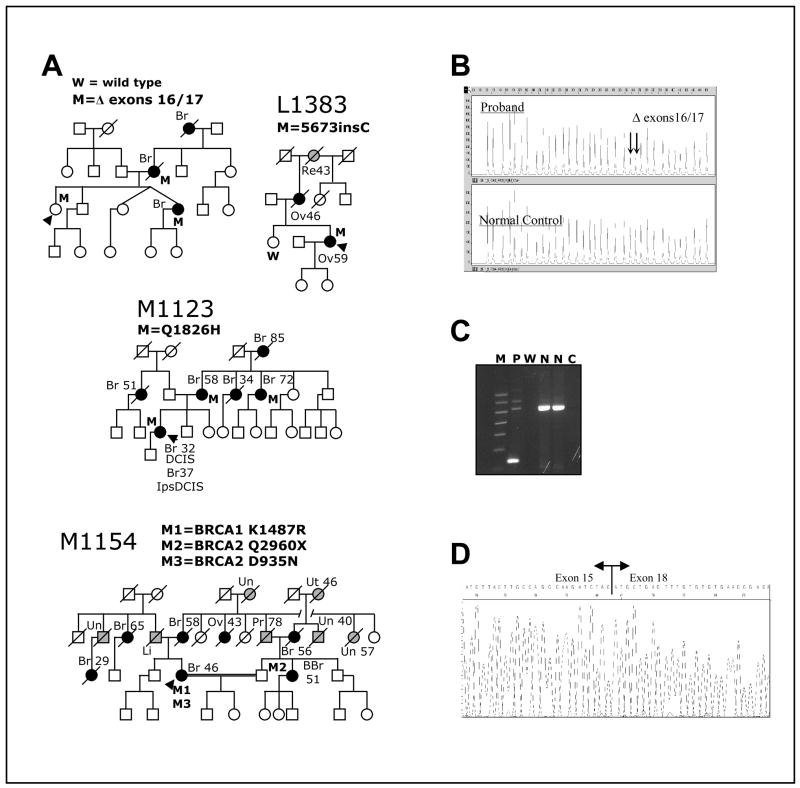
Fig. 2. Clinical data for BRCA1 variants.
A. Pedigrees of families carrying different variants. The presence (M) or absence (W) of the variants in the germ line of tested individuals is indicated. Proband is indicated by black arrowhead. Individuals affected by breast or ovarian cancer are denoted by a black circle or square and individuals affected by other cancers are denoted by a grey circle or square; site of tumor and age of diagnosis, when available is also indicated: Br, breast (individuals with two Br, reflect contralateral breast cancer); BBr, bilateral breast cancer; DCIS, ductal carcinoma in situ; Ips, ipsilateral; Li, liver; Ov, ovary; Pr, prostate; Re, rectal; Sk, skin; Un, unknown site; Ut, uterine. B. Multiplex Ligation-dependent Probe Amplification (MLPA) analysis of genomic DNA showing BRCA1 Δ exons 16/17. Note the halved intensity of the peaks indicated by arrows (corresponding to probes for exons 16 and 17, respectively) in the proband sample relative to the normal control specimen. It indicates genomic hemizygosity for BRCA1 Δ exons 16/17. C. Agarose gel of PCR products from cDNA with primers situated in exons 15 and 18 to amplify across the region containing the deletion shows BRCA1 deletion of exons 16 and 17 in proband’s RNA. M, molecular weight marker; P, Proband; W, water blank; N, normal control; C, RT blank. D. Sequencing of PCR amplicon from cDNA showing BRCA1 Δ exons 16/17 in proband’s RNA. Note that splicing of exon 15 to exon 18 preserves the reading frame.