Abstract
The telomere end-protection complex prevents the ends of linear eukaryotic chromosomes from degradation or inappropriate DNA repair. The homodimeric double-stranded DNA-binding protein, Trf1, is a component of this complex and is essential for mouse embryonic development. To define the requirement for Trf1 in somatic cells, we deleted Trf1 in chicken DT40 cells by gene targeting. Trf1-deficient cells proliferated as rapidly as control cells and showed telomeric localization of Trf2, Rap1, and Pot1. Telomeric G-strand overhang lengths were increased in late-passage Trf1-deficient cells, although telomere lengths were unaffected by Trf1 deficiency, as determined by denaturing Southern and quantitative FISH analysis. Although we observed some clonal variation in terminal telomere fragment lengths, this did not correlate with cellular Trf1 levels. Trf1 was not required for telomere seeding, indicating that de novo telomere formation can proceed without Trf1. The Pin2 isoform and a novel exon 4, 5–deleted isoform localized to telomeres in Trf1-deficient cells. Trf1-deficient cells were sensitive to DNA damage induced by ionizing radiation. Our data demonstrate that chicken DT40 B cells do not require Trf1 for functional telomere structure and suggest that Trf1 may have additional, nontelomeric roles involved in maintaining genome stability.
INTRODUCTION
The protein complex that binds to and stabilizes the telomeric DNA at the end of the linear eukaryotic chromosome, dubbed “shelterin” in a recent review (de Lange, 2005), comprises the double-stranded telomere repeat-binding proteins TRF1 and TRF2 and the single-stranded telomere-binding protein POT1, together with their interacting proteins TIN2, TPP1, and RAP1. Current models for shelterin organization have TIN2 linking TPP1/POT1 to TRF1 and TRF2/RAP1, with other possible subcomplexes (Liu et al., 2004; Ye et al., 2004). In addition to shelterin, components of the DNA damage response apparatus are required for telomere protection, notably DNA-PK (Bailey et al., 2004), Ku (Bianchi and de Lange, 1999; Hsu et al., 1999; d'Adda di Fagagna et al., 2001), and ATM (Karlseder et al., 2004).
The loss of shelterin function and the exposure of the telomere can have severe consequences for cells. Recent models invoke the recognition of the end of a linear chromosome as a DNA double-strand break (DSB) by the DNA repair apparatus as a crucial signal inducing senescence, with apoptosis as an alternative outcome (Blackburn, 2001; Karlseder et al., 2002; d'Adda di Fagagna et al., 2003; d'Adda di Fagagna et al., 2004). Loss of telomeric Trf2 caused by expression of a dominant-negative form or its genetic ablation results in deprotection of telomere ends and their being processed in a manner similar to DSBs, resulting in extensive chromosome fusions and anaphase bridging and causing apoptosis or cellular senescence (van Steensel et al., 1998; Takai et al., 2003; Celli and de Lange, 2005; Celli et al., 2006). These deprotected telomeres activate a DNA damage response to mediate these effects (Takai et al., 2003; Celli and de Lange, 2005), although recent data indicate that such recognition of telomere structures by ATM and ATR is a component of the normal cell cycle regulation of the telomere (Verdun et al., 2005; Verdun and Karlseder, 2006). Additional levels of regulation of the DNA damage response by Trf2 have been indicated by its binding to ATM (Karlseder et al., 2004), its being transiently phosphorylated after DNA damage (Tanaka et al., 2005) and its localization to nontelomeric DSBs (Bradshaw et al., 2005). Recent gen- targeting experiments in mouse and in chicken have confirmed a role for Pot1 in suppressing telomere DNA damage responses and endoreduplication, further emphasizing the importance of the DNA-binding constituents of shelterin in telomere and chromosome stability (Churikov et al., 2006; Hockemeyer et al., 2006; Wu et al., 2006).
Loss of Trf1 is lethal early in murine embryogenesis (Karlseder et al., 2003) and retards the proliferation of murine embryonic stem (ES) cells (Iwano et al., 2004). Trf1 is believed to modulate telomerase access to telomere ends, as overexpression of Trf1 causes telomere shortening and its inhibition leads to telomere elongation (van Steensel and de Lange, 1997; Smogorzewska et al., 2000). Trf1 has been described as an ATM interactor and phosphorylation target (Kishi et al., 2001; Kishi and Lu, 2002). It has also been described as being involved in mitotic regulation (Nakamura et al., 2001; Nakamura et al., 2002), so that the embryonic lethality in Trf1 nulls may result from telomere-independent defects. Its essential roles remain unclear, as yet. Here, we use gene targeting in the chicken DT40 cell line to explore the impact of Trf1 deficiency in somatic cells and describe the unexpected viability of these cells.
MATERIALS AND METHODS
Cloning and Cell Culture
Cloning and partial mapping of the chicken Terf1 locus was performed by restriction digest and Southern blotting and targeting vectors constructed by genomic cloning and PCR. Wild-type DT40 cells were cultured, and transfections and gene targeting were performed as described (Takata et al., 1998). Telomere seeding experiments were performed and analyzed as previously described (Farr et al., 1991). Reverse transcription was performed on total DT40 RNA using the Superscript First Strand kit (Invitrogen, Carlsbad, CA) and the cTrf1, cPin2, and cΔ4,5 Trf1 cDNAs were amplified with the oligonucleotides 5′-ATGTCGGAAGCGGGGAGG-3′ and 5′-TTATTTGATCTTGCACAG-3′ using LA Taq (Takara, Tokyo, Japan). cRap1 cDNA was amplified with the oligonucleotides 5′-ATGGCGGCACCCCCACGG-3′ and 5′-ACTTTTCCTAAACGCTAC-3′, and cPot1 cDNA was amplified using the oligonucleotides 5′-CCCAAGCTTGGGATGCCTGTTCAAGTGCTA-3′ and 5′-TACAACATCTTCAGCAAC-3′. The cDNA was then cloned into pCMV-3Tag-2 or pCMV-Tag2 (Stratagene. La Jolla, CA), and the constructs were sequenced. Ionizing radiation (IR) experiments were performed using a 137Cs source (Mainance Engineering, Waterlooville, United Kingdom). Clonogenic survival of wild-type and Terf1-null DT40 cells was determined as previously described (Takata et al., 1998). Cells were serially diluted from 1 × 105 to 1 × 103 cells/ml, plated in 5 ml methylcellulose media, and incubated for 1 h at 39.5°C before γ-irradiation. Colonies were counted 10 d after treatment, as described previously.
Antibodies
Primary antibodies used were anti-myc monoclonal 9E10, anti-FLAG monoclonal M2 (Sigma, St. Louis, MO), anti-γ-H2AX monoclonal JBW301 (Upstate Biotechnology, Lake Placid, NY) and anti-α-tubulin monoclonal B512 (Sigma). A polyclonal antiserum (93809) was raised in rabbit against the chicken Trf2 peptide RAPSPAERRKDLVRAPKRAET (Abcam, Cambridge, United Kingdom).
Microscopy
Immunofluorescence microscopy was performed using methanol-EGTA fixation as described (Dodson et al., 2004). For combined fluorescence in situ hybridization (FISH)/immunofluorescence microscopy, we used a modified version of published protocols (Kaminker et al., 2005). Cells were hypotonically swollen in 0.9% sodium citrate at 37°C, fixed in ice-cold methanol/acetic acid (3:1), and then dropped onto slides. They were then washed three times in 1× PBS and blocked in 0.1% BSA/PBS. The cells were incubated with the primary and secondary antibodies as described above. After three PBS washes, the slides were refixed in 4% paraformaldehyde for 2 min, washed three times in 1× PBS, and dehydrated in 70, 85, and 96% ethanol. The slides were air-dried and rehydrated in hybridization buffer (30% formamide, 0.1% Triton X-100, 0.3× SSC) containing a peptide nucleic acid (PNA) telomere probe (Panagene, Daejeon, Korea). The slides were denatured for 3 min at 80°C, incubated with the PNA probe for 30 min at room temperature, and then washed in 4× SSC, 0.05% Tween-20 for 5 min before counterstaining and mounting. Microscopy imaging was performed using an Olympus BX-51 microscope (Hamburg, Germany), 100× objective, NA 1.35 lens, driven by OpenLab software (version 3.1.4, Improvision, Coventry, United Kingdom). Deconvolved (Nearest Neighbor DCI, Improvision) images were saved as Adobe Photoshop CS files (version 8.0; San Jose, CA).
Telomerase Activity Assay
Protein extracts from DT40 cells were assayed for telomerase activity using the PCR-based telomeric repeat amplification protocol (TRAP) assay (Kim and Wu, 1997) as described (Faure et al., 2008).
Telomere Analyses and G-Strand Overhang Length Assays
To measure telomere fluorescence intensity in DT40 cells by quantitative FISH (Q-FISH), we hybridized metaphase chromosome preparations with the telomeric PNA oligonucleotide (CCCTAA)3 labeled with Cy3 (PE Biosystems, Foster City, CA) and acquired digital images as described (McIlrath et al., 2001). Telomere fluorescence intensity was analyzed using TFL-Telo Software provided by Drs. Peter Lansdorp and Steven S. S. Poon (Terry Fox Laboratory, Vancouver, BC, Canada). Telomere fluorescence was expressed in arbitrary units. To ensure reproducibility of our results, mouse lymphoma cell lines, LY-R and LY-S, with differing telomere lengths were used as internal standards. Because some chicken chromosomes contain interstitial telomeric sites, we used a fixed exposure time in all experiments to ensure accurate telomere fluorescence intensity measurement.
For native gel analysis of G-strand overhang length, 20 μg genomic DNA isolated using the Wizard Genomic DNA Purification kit (Promega, Madison, WI) was digested overnight with HinfI, MspI and HaeIII restriction enzymes and separated on a 0.7% agarose gel that was then dried and hybridized with a 32P-labeled (C3TA2)4 probe as described (Dionne and Wellinger, 1996). To control the specificity of the single-strand overhang signal, a probe specific for the C-strand was used, as well as digestion of the G-strand overhang with mung bean nuclease and exonuclease I was performed before restriction digestion of wild-type DNA (Wellinger et al., 1993). G-overhang signal was quantitated using ImageJ 1.34s software (http://rsb.info.nih.gov/ij/index.html/) and normalized using a representative band from the denatured gel TRF signal. For each experiment, telomere restriction fragment (TRF) length analysis by denaturing gel analysis was undertaken in parallel. Before drying, gels were washed in 2× SSC for 30 min at room temperature, denatured in 0.5 M NaOH, 1.5 M NaCl for 30 min, and neutralized in NaCl 1.5 M, Tris-Cl 0.5 M, pH 7.5, for 30 min. Signals were detected on an FLA-5100 PhosphorImager (Fuji, Tokyo, Japan). For TRF analysis, the scanned gels were analyzed as previously described for chicken telomeres (Wei et al., 2002), with the individual lanes being divided into 40 regions and the signal and fragment size determined using comparison with size markers in Image Quant (Molecular Dynamics, Sunnyvale, CA). Calculations of mean telomere length were performed using a previously published formula (Wei et al., 2002).
RESULTS
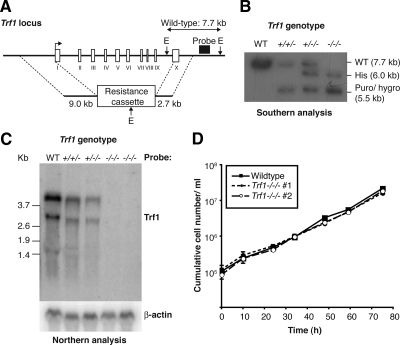
We have used gene targeting to disrupt Trf1 in the chicken DT40 cell line. Antibiotic cassettes were cloned between homology regions upstream and downstream of the Trf1 coding sequence (Figure 1A) and the resulting plasmids were used to target sequentially the Trf1 alleles in DT40 cells. Because Trf1 is located on chicken chromosome 2, which is usually trisomic in DT40 cells, this necessitated three rounds of gene targeting (Figure 1B). Several viable clones in which the wild-type Trf1 locus had been disrupted were independently derived. Northern blot analysis was used to confirm the genetic ablation of Trf1. As shown in Figure 1C, the loss of each allele of Trf1 resulted in a decline in the Trf1 transcript until no message was detected in either of the two null clones analyzed. These data demonstrate that there was a complete disruption of Trf1 after gene targeting. Next, we monitored the proliferation of the Trf1-deficient cells. We observed no difference in population doubling time between wild-type and Trf1 null clones (Figure 1D and data not shown). Analysis of four additional Trf1-deficient clones generated in separate experiments gave the same results (data not shown). Karyotype analysis of the Trf1 null clones revealed one to be disomic for chromosome 2 while retaining three Trf1 alleles (as detected by Southern analysis). This was not a general phenomenon associated with Trf1 targeting and has been described as occurring spontaneously in DT40 cultures (Chang and Delany, 2004). No evidence for macrochromosome fusions was seen in experiments counting 50 metaphases of two separate Trf1-deficient clones (data not shown). These findings demonstrate that Trf1 is not required for proliferation of DT40 cells.
Figure 1.
Gene targeting of Trf1 and preliminary phenotypic analysis. (A) Diagrammatic representation of the chicken Trf1 locus and gene targeting strategy. Exons are shown by white boxes and labeled with Roman numerals. E, relevant EcoRI sites. (B) Southern blot analysis of sequential Trf1 targeting steps in clones of the indicated genotypes. Expected sizes of the wild-type and targeted alleles are indicated at right. (C) Northern blot analysis of clones of the indicated genotypes hybridized with the full-length cDNA probes shown at right. Size markers at left are in kb. (D) Proliferation analysis of wild-type and two Trf1−/−/− clones. Data points are the mean ± SD of three separate experiments.
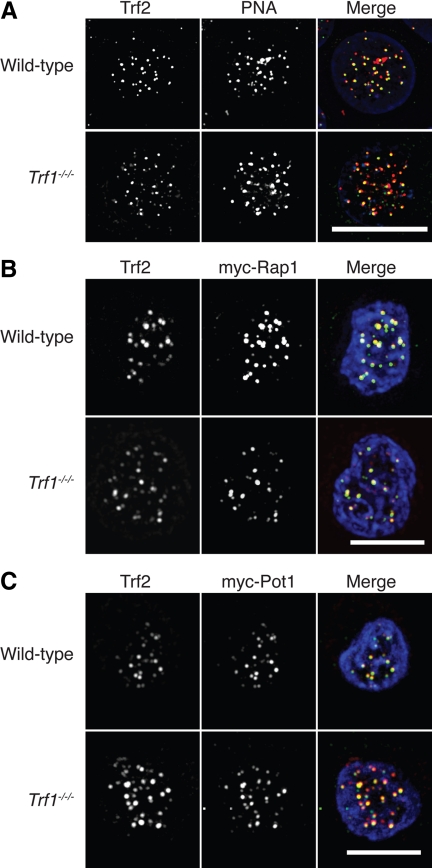
Because Trf1 is a key component of the telomere end-protection complex, we used microscopy to test whether Trf1 deletion impacts on the ability of other shelterin components to localize to telomere structures. We observed consistent colocalization of a Trf2 signal detected by immunofluorescence microscopy with the signal from a PNA probe hybridized to the telomere repeat sequence in wild-type and Trf1-deficient DT40 cells (Figure 2A), suggesting that Trf2 localizes normally to telomeres in the absence of Trf1. We also found robust colocalization of myc-Rap1 and myc-Pot1 with the Trf2 signal in wild-type and Trf1 null cells (Figure 2, B and C), showing that the localization of these components to telomeres does not require Trf1.
Figure 2.
Microscopy analysis of shelterin components in Trf1-deficient cells. (A) Cells of the indicated genotype were fixed and subjected to in situ hybridization with a PNA anti-telomere probe (red) then stained with antibodies to Trf2 (green) and counterstained with DAPI (blue). Cells of the indicated genotype were transiently transfected with expression vectors for myc-Rap1 (B) or myc-Pot1 (C) and then fixed and stained with antibodies to myc (red) and to Trf2 (green) and counterstained with DAPI (blue). Scale bars, 10 μm.
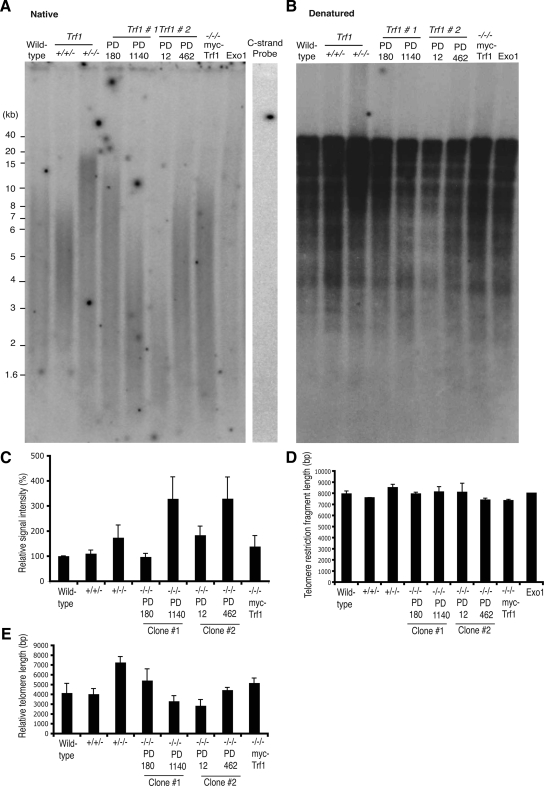
We next examined the impact of Trf1 deficiency on telomere length and on the G-strand overhang. Analysis of genomic DNA hybridized with a G-strand–specific probe under native conditions showed an increase in G-overhang signal in late passage Trf1-deficient cells when compared with wild-type, Trf1 heterozygote and early passage Trf1-deficient cells, as measured by intensity of the signal (Figure 3, A and C). These data suggest that G-overhang length progressively increases over multiple passages in the absence of Trf1, whereas overexpression of myc-Trf1 maintained overhang signal at wild-type levels (Figure 3, A and C; Supplemental Figure S1). We controlled for specificity of the G-strand probe by hybridizing a C-strand probe and by exonuclease digestion of any single-stranded sequence. No signal was observed in either of these negative control experiments (Figure 3A). Parallel analysis of genomic DNA with hybridization performed under denaturing conditions indicated no impact of Trf1 deficiency on telomere length (Figure 3, B and D). Although this assay also detects chicken interstitial sequences and is not an optimal measure of telomere length (Venkatesan and Price, 1998), it can detect severe reductions in telomere length, as in Ku70-deficient TR+/− DT40 cells (Faure et al., 2008). When we measured the length of the TRF fragments in the native gel, we noted a distinct clonal variation in telomere length that occurred over time (Figure 3E). Other DT40 clones examined by this assay have also shown some clonal variation in terminal TRF length (Wei et al., 2002; Faure et al., 2008), so that, in the absence of a clear trend resulting from Trf1 deficiency, we cannot conclude that this is a significant effect of Trf1 loss.
Figure 3.
Telomere maintenance and telomerase activity in Trf1-deficient cells. Autoradiogram showing native (A) and denatured (B) in-gel hybridizations of (TA2C3)4 G-strand probe to MspI-, HinfI-, and HaeIII-digested genomic DNA from the indicated cell lines at the population doublings (PD) indicated. As controls, wild-type samples were treated with ExoI or mung bean nuclease (MBN) before restriction digestion. Wild-type DNA was probed with a C-strand probe, which has sequence complementary to the G-strand probe. Trf1−/−/− myc-Trf1–expressing cells were passaged for ∼800–900 PD. (C) Histogram showing relative G-overhang signals. The signal in each lane was quantified with a phosphoimager and normalized against the signal detected in a standardized region in the denatured gel. Data represent the mean values + SEM of three independent experiments. (D) Histogram showing average telomere length for the indicated clones, as determined by analysis of the denaturing gel. Signals were measured using Image Quant, with each lane being divided into 40 boxes, and the weighted mean telomere length was calculated using the formula previously described (Wei et al., 2002). Data shown are from three independent experiments. (E) Histogram showing average telomere length for the indicated clones, as determined by analysis of the nondenaturing gel using the same approach as in D.
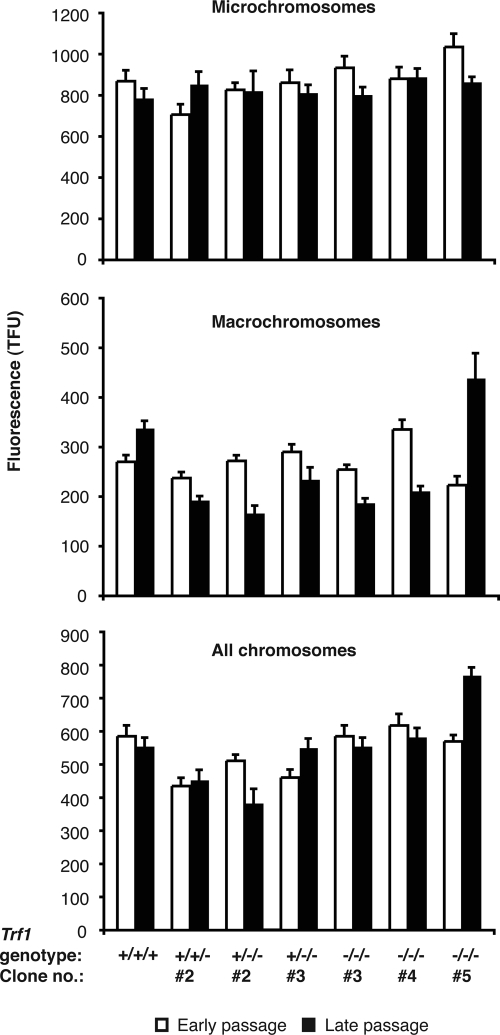
No effect of Trf1 deficiency was seen in in vitro analyses of telomerase activity (Supplemental Figure S2), consistent with Trf1 acting in cis to regulate telomerase activity (van Steensel and de Lange, 1997). The absence of a clear impact of Trf1 disruption on telomere length was unexpected, so we used Q-FISH analysis to determine telomere lengths in Trf1-deficient cells. A series of clones with varying Trf1 copy number were prepared for FISH and hybridized with a telomere-specific PNA probe. Metaphase spreads were stained with DAPI and analysis of telomere signals in macro- and microchromosomes was performed on digital images of 200–800 metaphase per clone analyzed. Overall, telomeres from macrochromosomes ranged in size from 17 to 43 kb, whereas those of the microchromosomes were 70 kb–1Mb in size [100 telomere fluorescence units (TFU) = 10 kb of (TTAGGG)n repeats]. As shown in Figure 4, all clones showed some fluctuation around a mean telomere length, but we saw no correlation between Trf1 copy number and telomere length. One clone did demonstrate an increase in macrochromosome telomere length with increasing passage number, but this was not a general trend. Thus, although the increase in G-strand overhang length observed in Trf1-deficient cells would be consistent with a role as a negative regulator in telomere maintenance, both molecular and Q-FISH analyses of terminal (TTAGGG)n repeat array length suggest that Trf1 is not required for telomere length homeostasis in DT40. Further work will be required to determine the mechanism by which Trf1 influences the G-strand overhang in this system.
Figure 4.
Q-FISH analysis of (TTAGGG)n fluorescence of micro- and macro-chromosomes in wild-type DT40 and Trf1 mutant cell lines. A Cy3-conjugated PNA probe complementary to the (TTAGGG)n repeat array was hybridized to DAPI-stained metaphase spreads of DT40 cells of the indicated genotypes. Fluorescence intensities were measured at early passage (within 30 PD of the clonal cell line being established) and again at late passage (≈100 PDs later for all lines with the exception of one of the Trf1−/−/− mutants, which was resampled after only a further ∼50 PDs). Data were collected from as many chromosome ends as possible per metaphase spread and from between 200 and 800 metaphases for each cell line examined. (A) Fluorescence data for microchromosomes only, (B) for macrochromosomes only, and (C) pooled data for both micro- and macro-chromosomes. Values represent the mean ± SEM.
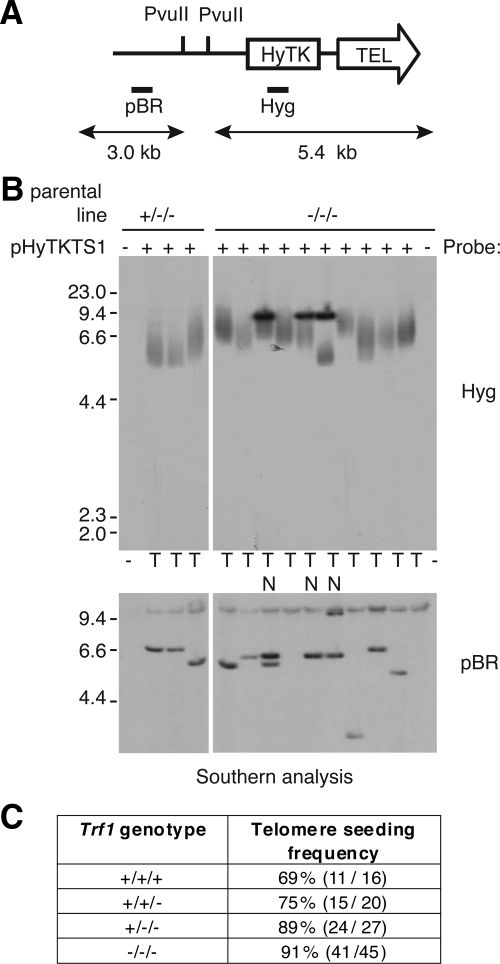
Introduced DNA fragments containing telomere repeat sequence can induce de novo formation of telomeres at their sites of integration into the chromosome, in a process called telomere seeding (Farr et al., 1991). Correlative experiments having implicated Trf1 in this activity (Okabe et al., 2000), we tested whether Trf1 deficiency altered telomere seeding in DT40 cells. After transfection of the seeding construct shown in Figure 5A, Southern analysis of genomic DNA after a low number of population doublings was used to determine whether the telomere region of the construct had expanded, as determined by the presence of a smear when hybridized with the resistance cassette as probe (Figure 4B). No reduction of telomere seeding frequency was observed in the absence of Trf1 (Figure 5, B and C). Because DT40 has a high rate of targeted integration of homologous DNA, we investigated the possibility that the seeding constructs had integrated into existing telomeres. To recover genomic DNA proximal to de novo seeded telomeres, we performed plasmid rescue on a number of clones with single, telomeric integrations of the seeding construct. Flanking DNA was successfully recovered from four transfectants (two Trf1−/−/− and two wild-type DT40 clones), and its origin was confirmed by Southern blotting. Sequencing of several hundred base pairs from both ends of the rescued DNAs from sites in the pBR322 plasmid backbone, revealed no (TTAGGG)n repeat or its variants (data not shown). This is consistent with the telomeric integrations observed in the DT40-derived lines due to genuine telomere seeding rather than targeted integration at existing telomeres. The fact that de novo telomere seeding events can occur both in wild-type DT40 and in the Trf1 null background suggest that Trf1 is not required for either telomere maintenance or generation in this system.
Figure 5.
Analysis of transfectants for de novo telomere seeding. (A) Schematic of the seeding construct pHyTKTS1, linearized with NotI. The PvuII sites and probe DNAs (Hyg and pBR) are indicated. If upon integration a telomere is generated de novo the terminal PvuII restriction fragment will be a heterogeneous smear of ≥5.4 kb. (B) Southern blot analysis of PvuII-digested DNA from pHyTKTS1 transfectants derived from Trf1+/−/− and Trf1−/−/− cell lines. The outermost lanes contain DNA from the two mutant cell lines before transfection with the seeding construct. In the top panel the filter has been probed using Hyg to detect potential terminal restriction fragments, whereas in the bottom panel the filter has been stripped and reprobed using the plasmid backbone to confirm the integrity of the genomic DNA. Clones with an internally located construct are marked N, and those with a seeded telomere are marked T. (C) Table summarizing telomere seeding frequency in cell lines with different numbers of Trf1 alleles (percentage, plus number of transfectants with a de novo telomere out of the total analyzed).
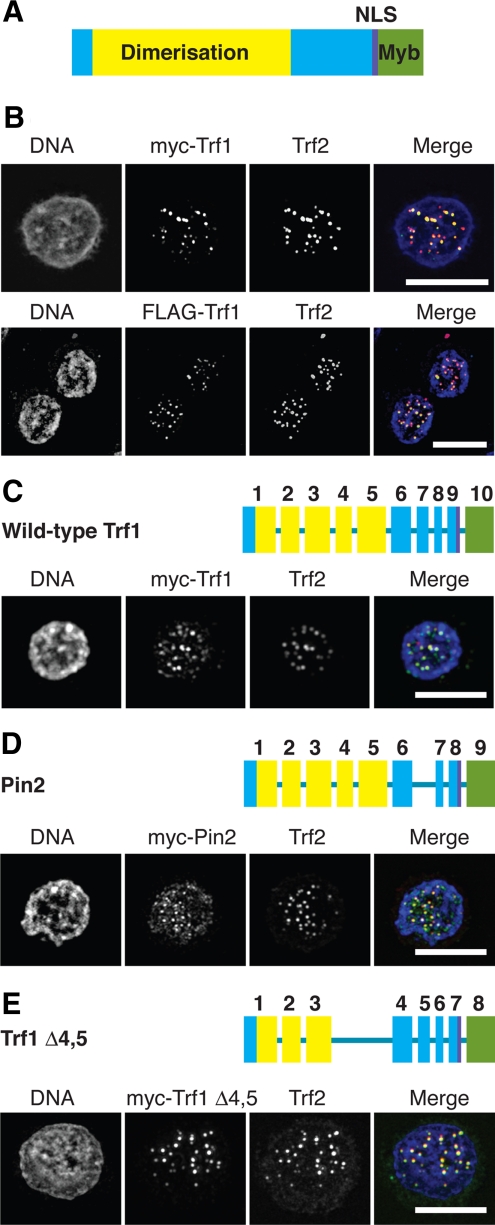
We next sought to exploit the Trf1-deficient cells as a vehicle to explore the roles of Trf1 splice variants, because a role in mitosis had been suggested for Trf1 and an abundant splice variant, Pin2 (Shen et al., 1997; Nakamura et al., 2001, 2002). We used RT-PCR to clone chicken Pin2. During this cloning, we also identified a novel variant of Trf1 in which exons 4 and 5 are absent, Trf1Δ4,5 (Accession number EU688999). Next, Trf1, Pin2, and Trf1Δ4,5 were expressed as myc-tagged fusions in Trf1−/−/−cells and monitored for colocalization with Trf2 at telomeres. Fusion proteins of myc- and FLAG-tagged full-length Trf1 both colocalized with Trf2 at telomeres in DT40 cells (Figure 6, A and B). As shown in Figure 6, C–E, myc-tagged full-length Trf1, Pin2, and Trf1Δ4,5 colocalized with Trf2 at telomeres in the absence of any endogenous Trf1. These data show that the minimal region of chicken Trf1 required for dimerization and telomere localization does not include the protein sequence encoded by exons 4 and 5. We saw no localization of myc-Pin2 with the mitotic spindle in chicken DT40 cells (data not shown). Although these findings do not exclude a role for Trf1/Pin2 in mitosis, the absence of any obvious defect in cell cycle progression suggests that the loss or reintroduction of Trf1 or Pin2 does not significantly affect mitosis.
Figure 6.
Microscopy analysis of splice variants of Trf1. (A) Domain structure of chicken Trf1, based on published data for the human orthologue. (B) Wild-type DT40 cells were transfected with expression vectors for myc-Trf1 or FLAG-Trf1 and then fixed and stained with antibodies to myc or FLAG, respectively (green), and to Trf2 (red) and counterstained with DAPI (blue). Scale bars, 10 μm. Trf1−/−/− DT40 cells were transfected with expression vectors for myc-Trf1 (C), myc-Pin2 (D), or myc-Trf1Δ4,5 (E) and then fixed and stained with antibodies to myc (green) and Trf2 (red) and counterstained with DAPI (blue). Exon diagrams use the same color scheme as A and indicate the exons deleted in the splice variants of Trf1. Scale bars, 10 μm.
Whereas performing transfections of various expression constructs into Trf1−/−/−cells, we noted that electroporation of Trf1-deficient cells caused a sustained DNA damage response, as measured by H2AX phosphorylation (Supplemental Figure S3). Therefore, we examined the role of Trf1 in the cellular response to induced DNA damage by subjecting wild-type and Trf1−/−/− cells to IR. As shown in Figure 7, A and B, Trf1-deficient cells were more sensitive to IR than wild-type controls. Analysis of the time course of this DNA damage response by immunoblot of γ-H2AX confirmed this sensitivity, with the Trf1-deficient clones having a greatly extended period during which a γ-H2AX signal was detected (Figure 7C). We saw no background H2AX phosphorylation and no telomere-localizing DNA damage foci in Trf1-deficient cells (Figure 7C and data not shown), indicating that Trf1 deficiency does not activate a constitutive DNA damage response in DT40 cells. The extent to which Trf1 contributes to IR sensitivity is not clear from our experiments, however, because we also observed IR sensitivity in Trf1+/+/− and Trf +/−/− cells and this phenotype was not rescued by expression of myc-Trf1 in Trf1-deficient cells (data not shown). On the other hand, the detection of this IR sensitivity in multiple Trf1 mutant clones suggests that DT40 cells with reduced levels of Trf1 may be particularly sensitive to DNA DSBs.
Figure 7.
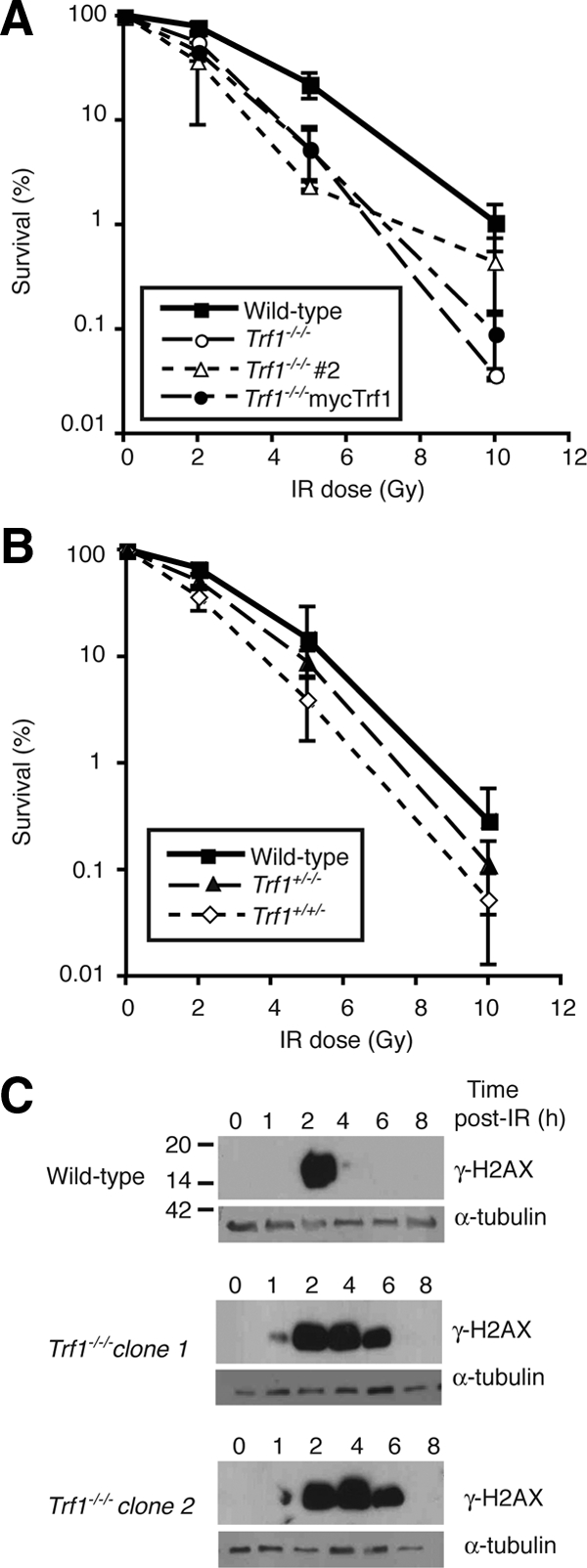
Defective responses to ionizing radiation in Trf1-deficient cells. (A and B) Clonogenic survival of cells of the indicated genotype after treatment with different doses of γ-irradiation. Data points show mean ± SD of three separate triplicate experiments. (C) Immunoblot analysis of H2AX phosphorylation in cells of the indicated genotype at the indicated times after 10 Gy γ-irradiation. α-Tubulin is used as a loading control, and these blots are representative of three separate experiments. Indicative size markers are shown at left in kDa.
DISCUSSION
We have described the unexpected viability of chicken DT40 cells that lack Trf1. Work in the mouse system has shown that Trf1 is required for embryonic viability and for ES cell proliferation (Karlseder et al., 2003; Iwano et al., 2004). The essential function of Trf1 is not clear from analysis of these models, however. A reduction of telomeric Tin2, Trf2, Tpp1, and Pot1 was noted in conditionally Trf1-deficient ES cells after Trf1 depletion (Iwano et al., 2004; Okamoto et al., 2008). This loss of telomeric shelterin components was accompanied by increased chromosome abnormalities and telomere fusions (Iwano et al., 2004), which were suppressed by the enforced localization of shelterin subunits to Trf1-deficient telomeres (Okamoto et al., 2008). However, analysis of metaphase spreads from Trf1-deficient blastocysts did not reveal uncapped telomere fusions or chromosome instability, with the caveat that apoptosis may have removed highly aberrant cells from the population before analysis (Karlseder et al., 2003). Partial depletion of TRF1 by RNA interference in human tumor cells also reduced telomeric TRF2 and hRap1 (Ye et al., 2004), consistent with a cooperative role for TRF1 in establishing the telomere end-protection complex. Nevertheless, small interfering RNA (siRNA)-mediated loss of TRF1 in another human cell line did not cause gross toxicity or disrupt the cell cycle profile within a 72-h experimental period (Deng et al., 2003).
Clearly, our data are not consistent with findings made in mammalian model systems. We envisage four potential explanations that focus on the telomeric roles of Trf1. First, that the complete deletion of the Trf1 locus in chicken cells causes a different phenotype to the targeting of murine exon 1 because of the expression of some aberrant form of Trf1 in homozygous exon 1–targeted cells. However, there was no evidence of a truncated Trf1 transcript in Trf1 heterozygote MEFs (Karlseder et al., 2003) and no abnormal Trf1 gene products were described in any of the articles on the targeting of mouse Trf1 (Karlseder et al., 2003; Iwano et al., 2004; Okamoto et al., 2008). Disruption of another, unknown mouse gene that is nonsyntenic with its chicken orthologue is a formal possibility, but the rescue of the Trf1-deficient phenotypes by stable expression of FLAG-mTrf1 argues strongly against this notion (Okamoto et al., 2008).
A second possibility is that somatic cells respond to telomere dysfunction in a manner distinct from mouse embryonic cells. The absence of p53 from DT40 cells is a potentially important source of variation in the phenotype. However, p53 deletion extended the embryonic viability of mouse Trf1 knockouts only briefly, showing that the lethality of Trf1 deficiency in mouse is not p53-dependent (Karlseder et al., 2003). Telomere dysfunction induces a similar response in DT40 cells and mouse cells, with deletion of the single Pot1 isoform in chicken and of the two isoforms in mouse causing increased G-strand overhang length, cell cycle arrest and limited numbers of chromosome fusions (Churikov et al., 2006; Hockemeyer et al., 2006). Telomere dysfunction caused by disruption of one allele of Terc in DT40 cells also caused cell cycle arrest (Faure et al., 2008). Therefore, it seems unlikely that DT40 cells are unable to recognize or respond to telomere abnormalities.
A third hypothesis is that there is an as-yet undescribed functional homologue of Trf1 in chickens. Mice have two Pot1 paralogues, as opposed to the single gene seen in humans and chickens, suggesting that some interspecies variation in shelterin makeup is possible (Churikov et al., 2006; Hockemeyer et al., 2006; Wu et al., 2006). However, recent identification of Tbf1p, a novel telomere repeat binding factor, in Schizosaccharomyces pombe, in which only Taz1p had been described as a TRF1/TRF2 orthologue, indicates that two TRF1/TRF2 orthologues are conserved between fission yeast and mammals (Pitt et al., 2008). In addition, despite screening by Southern blot, Northern blot, and expressed sequence tag searching, we have no evidence for more than a single Trf1 gene in chicken.
Our fourth potential explanation is that the composition of the shelterin complex found in chicken may differ from that in mammals. Chicken Trf1 is a slightly smaller protein than its mammalian homologues, with the N-terminal domain showing little similarity either in length (19 amino acids compared with 68 and 55 in human and mouse Trf1, respectively) or amino acid sequence identity (10–17%; De Rycker et al., 2003; Crumet et al., 2006). The localization of Trf2, Rap1, and Pot1 to Trf1-deficient telomeres and the absence of telomere dysfunction-induced damage foci (TIFs) in Trf1−/−/−cells are clear evidence that a functional telomere end-protection complex can be made in the absence of Trf1 in DT40 cells. One possibility is that Trf2 can substitute for Trf1. Although the pronounced response of Trf1-deficient cells to transient transfection has impeded our testing this hypothesis by siRNA or overexpression of dominant- negative Trf2 isoforms in the DT40 system, it has been explored directly in murine ES cells. Although enforced telomeric localization of Tin2 in Trf1-deficient mouse ES cells restored Tpp1, Pot1a, and Pot1b to telomere foci, telomeric Trf2 localization in the absence of Trf1 was not sufficient to do so (Okamoto et al., 2008). However, enforced telomeric localization of Trf2 or Tin2 rescued both the proliferation defect and the TIF phenotype (Okamoto et al., 2008). This observation raises the possibility that the lethality of Trf1 deficiency in mouse embryonic cells is due not only to the disruption of shelterin, but also to other activities of Trf1 in genome maintenance that may be required for mouse embryonic, but not DT40, cell viability. Analysis of the impact of TRF1 deficiency on mammalian somatic cells will clarify which of these hypotheses is correct.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carolyn Price for advice and discussion the following for their help in the early stages of this project: Johanna Abbott, Patrick Hextall, Jonathan Konrad, Helena Seth-White, and Suzanne Whittle. Work in C.M.'s lab was supported by a Science Foundation Ireland Investigator award. K.M.B. was supported by an Medical Research Council studentship and J.L.S. by the Cambridge European Trust, a Trinity Hall Bursary, a Sackler Award, and a Robert Gardiner Memorial Award.
Abbreviations used:
- DSB
DNA double-strand break
- ES cell
embryonic stem cell
- MEF
murine embryonic fibroblast
- PNA
peptide nucleic acid.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-10-1019) on March 25, 2009.
REFERENCES
- Bailey S. M., Brenneman M. A., Halbrook J., Nickoloff J. A., Ullrich R. L., Goodwin E. H. The kinase activity of DNA-PK is required to protect mammalian telomeres. DNA Repair. 2004;3:225–233. doi: 10.1016/j.dnarep.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Bianchi A., de Lange T. Ku binds telomeric DNA in vitro. J. Biol. Chem. 1999;274:21223–21227. doi: 10.1074/jbc.274.30.21223. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- Bradshaw P. S., Stavropoulos D. J., Meyn M. S. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat. Genet. 2005;37:193–197. doi: 10.1038/ng1506. [DOI] [PubMed] [Google Scholar]
- Celli G. B., de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Celli G. B., Denchi E. L., de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- Chang H., Delany M. E. Karyotype stability of the DT40 chicken B cell line: macrochromosome variation and cytogenetic mosaicism. Chromosome Res. 2004;12:299–307. doi: 10.1023/b:chro.0000021947.17128.96. [DOI] [PubMed] [Google Scholar]
- Churikov D., Wei C., Price C. M. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol. Cell. Biol. 2006;26:6971–6982. doi: 10.1128/MCB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumet N., Carlson R. L., Drutman S. B., Shampay J. A truncated acidic domain in Xenopus TRF1. Gene. 2006;369:20–26. doi: 10.1016/j.gene.2005.10.006. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Hande M. P., Tong W. M., Roth D., Lansdorp P. M., Wang Z. Q., Jackson S. P. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Reaper P. M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N. P., Jackson S. P. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Teo S. H., Jackson S. P. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- De Rycker M., Venkatesan R. N., Wei C., Price C. M. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem. J. 2003;372:87–96. doi: 10.1042/BJ20021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Atanasiu C., Burg J. S., Broccoli D., Lieberman P. M. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J. Virol. 2003;77:11992–12001. doi: 10.1128/JVI.77.22.11992-12001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne I., Wellinger R. J. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson H., Bourke E., Jeffers L. J., Vagnarelli P., Sonoda E., Takeda S., Earnshaw W. C., Merdes A., Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr C., Fantes J., Goodfellow P., Cooke H. Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA. 1991;88:7006–7010. doi: 10.1073/pnas.88.16.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure V., Wenner T., Cooley C., Bourke E., Farr C. J., Takeda S., Morrison C. G. Ku70 prevents genome instability resulting from heterozygosity of the telomerase RNA component in a vertebrate tumour line. DNA Repair. 2008;7:713–724. doi: 10.1016/j.dnarep.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Daniels J. P., Takai H., de Lange T. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Hsu H. L., Gilley D., Blackburn E. H., Chen D. J. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano T., Tachibana M., Reth M., Shinkai Y. Importance of TRF1 for functional telomere structure. J. Biol. Chem. 2004;279:1442–1448. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- Kaminker P., Plachot C., Kim S. H., Chung P., Crippen D., Petersen O. W., Bissell M. J., Campisi J., Lelievre S. A. Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2. J. Cell Sci. 2005;118:1321–1330. doi: 10.1242/jcs.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J., Hoke K., Mirzoeva O. K., Bakkenist C., Kastan M. B., Petrini J. H., de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J., Kachatrian L., Takai H., Mercer K., Hingorani S., Jacks T., de Lange T. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 2003;23:6533–6541. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J., Smogorzewska A., de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- Kim N. W., Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP) Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S., Lu K. P. A critical role for Pin2/TRF1 in ATM-dependent regulation. Inhibition of Pin2/TRF1 function complements telomere shortening, radiosensitivity, and the G(2)/M checkpoint defect of ataxia-telangiectasia cells. J. Biol. Chem. 2002;277:7420–7429. doi: 10.1074/jbc.M111365200. [DOI] [PubMed] [Google Scholar]
- Kishi S., Zhou X. Z., Ziv Y., Khoo C., Hill D. E., Shiloh Y., Lu K. P. Telomeric protein Pin2/TRF1 as an important ATM target in response to double strand DNA breaks. J. Biol. Chem. 2001;276:29282–29291. doi: 10.1074/jbc.M011534200. [DOI] [PubMed] [Google Scholar]
- Liu D., O'Connor M. S., Qin J., Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- McIlrath J., et al. Telomere length abnormalities in mammalian radiosensitive cells. Cancer Res. 2001;61:912–915. [PubMed] [Google Scholar]
- Nakamura M., Zhou X. Z., Kishi S., Kosugi I., Tsutsui Y., Lu K. P. A specific interaction between the telomeric protein Pin2/TRF1 and the mitotic spindle. Curr. Biol. 2001;11:1512–1516. doi: 10.1016/s0960-9822(01)00456-0. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Zhou X. Z., Kishi S., Lu K. P. Involvement of the telomeric protein Pin2/TRF1 in the regulation of the mitotic spindle. FEBS Lett. 2002;514:193–198. doi: 10.1016/s0014-5793(02)02363-3. [DOI] [PubMed] [Google Scholar]
- Okabe J., Eguchi A., Masago A., Hayakawa T., Nakanishi M. TRF1 is a critical trans-acting factor required for de novo telomere formation in human cells. Hum. Mol. Genet. 2000;9:2639–2650. doi: 10.1093/hmg/9.18.2639. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Iwano T., Tachibana M., Shinkai Y. Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J. Biol. Chem. 2008;283:23981–23988. doi: 10.1074/jbc.M802395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt C. W., Valente L. P., Rhodes D., Simonsson T. Identification and characterization of an essential telomeric repeat binding factor in fission yeast. J. Biol. Chem. 2008;283:2693–2701. doi: 10.1074/jbc.M708784200. [DOI] [PubMed] [Google Scholar]
- Shen M., Haggblom C., Vogt M., Hunter T., Lu K. P. Characterization and cell cycle regulation of the related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis. Proc. Natl. Acad. Sci. USA. 1997;94:13618–13623. doi: 10.1073/pnas.94.25.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M. R., Schnapp G., de Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Takata M., Sasaki M. S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Mendonca M. S., Bradshaw P. S., Hoelz D. J., Malkas L. H., Meyn M. S., Gilley D. DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proc. Natl. Acad. Sci. USA. 2005;102:15539–15544. doi: 10.1073/pnas.0507915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Venkatesan R. N., Price C. Telomerase expression in chickens: constitutive activity in somatic tissues and down-regulation in culture. Proc. Natl. Acad. Sci. USA. 1998;95:14763–14768. doi: 10.1073/pnas.95.25.14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun R. E., Crabbe L., Haggblom C., Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Verdun R. E., Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Wei C., Skopp R., Takata M., Takeda S., Price C. M. Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res. 2002;30:2862–2870. doi: 10.1093/nar/gkf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger R. J., Wolf A. J., Zakian V. A. Structural and temporal analysis of telomere replication in yeast. Cold Spring Harb. Symp. Quant. Biol. 1993;58:725–732. doi: 10.1101/sqb.1993.058.01.080. [DOI] [PubMed] [Google Scholar]
- Wu L., et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Ye J. Z., Donigian J. R., van Overbeek M., Loayza D., Luo Y., Krutchinsky A. N., Chait B. T., de Lange T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.