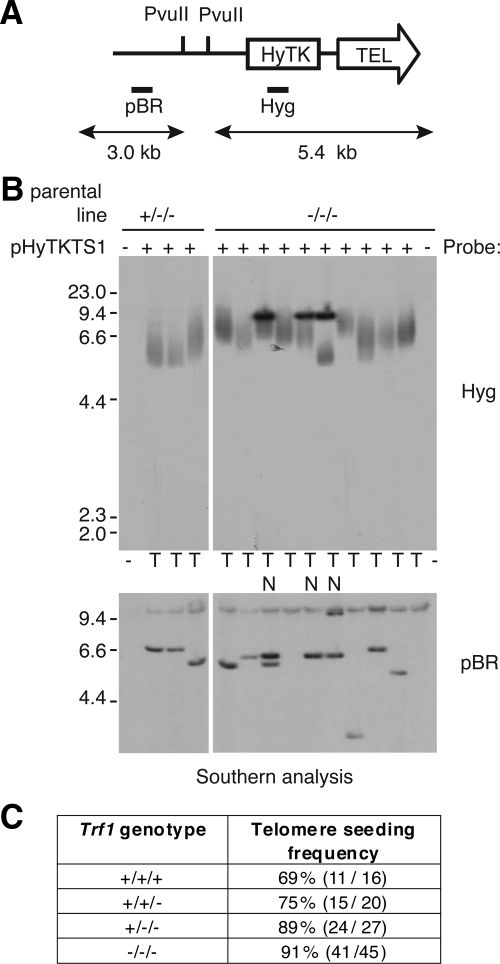
Figure 5.
Analysis of transfectants for de novo telomere seeding. (A) Schematic of the seeding construct pHyTKTS1, linearized with NotI. The PvuII sites and probe DNAs (Hyg and pBR) are indicated. If upon integration a telomere is generated de novo the terminal PvuII restriction fragment will be a heterogeneous smear of ≥5.4 kb. (B) Southern blot analysis of PvuII-digested DNA from pHyTKTS1 transfectants derived from Trf1+/−/− and Trf1−/−/− cell lines. The outermost lanes contain DNA from the two mutant cell lines before transfection with the seeding construct. In the top panel the filter has been probed using Hyg to detect potential terminal restriction fragments, whereas in the bottom panel the filter has been stripped and reprobed using the plasmid backbone to confirm the integrity of the genomic DNA. Clones with an internally located construct are marked N, and those with a seeded telomere are marked T. (C) Table summarizing telomere seeding frequency in cell lines with different numbers of Trf1 alleles (percentage, plus number of transfectants with a de novo telomere out of the total analyzed).