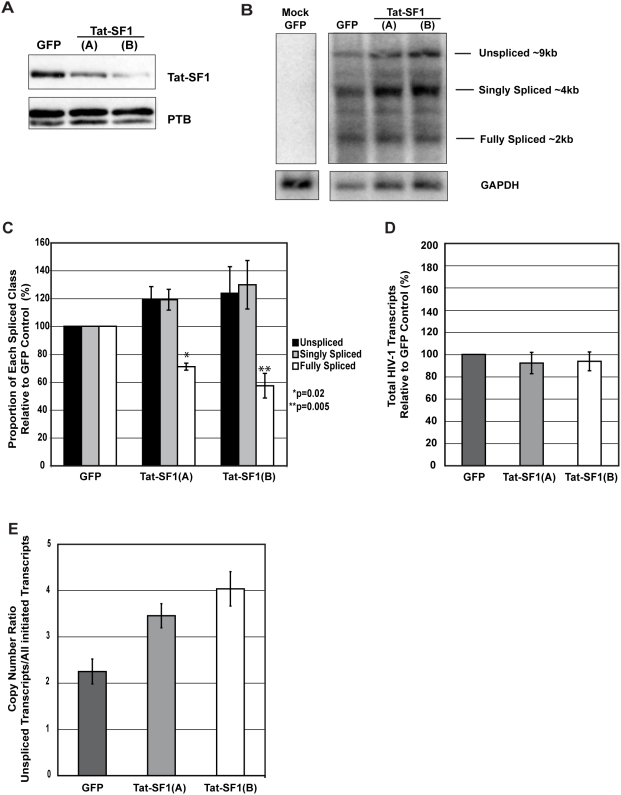
Figure 5. Tat-SF1 maintains the levels of unspliced and spliced HIV-1 RNAs.
(A) Western blot analysis confirming knockdown in T-Rex-293 cells. (B) Representative Northern blot analysis of HIV-1 RNA classes. T-Rex-293 cells were transfected with pSG3ΔEnv 72 hours after tetracycline induction. At 48 hours post-transfection, total RNA was isolated for electrophoresis and Northern blotting. A DNA probe specific to the HIV-1 LTR detected the ∼9 kb unspliced, ∼4 kb singly spliced, and ∼2 kb fully spliced RNAs. Lane 1 contains RNA from mock-transfected GFP control cells, lane 2 from transfected GFP control cells, and lanes 3 and 4 from transfected Tat-SF1 shRNA cells. The lower panel shows the same membrane, stripped and reprobed for GAPDH. (C) Tat-SF1 depletion alters the levels of HIV-1 RNA classes. Values are reported as the mean proportion of each RNA class, relative to the GFP control cells from three independent Northern blot experiments. Error bars represent standard error. Statistically significant differences between GFP control and Tat-SF1 knockdown conditions are indicated with asterisks. (D) Tat-SF1 depletion does not alter total HIV-1 RNA levels. Levels of the 3 RNA classes quantified from triplicate Northern blots were totaled and normalized to GAPDH levels. Values are reported as the means relative to the GFP control cells from three independent experiments. Error bars represent standard error. (E) Tat-SF1 depletion results in an increase in unspliced HIV-1 transcripts. qRT-PCR was performed on the same RNA samples used for Northern blot experiments. The medians of triplicate amplifications (both unspliced products and all initiated HIV-1 transcripts) were calculated and means of unspliced transcripts/all initiated HIV-1 transcripts ratios from triplicate samples are reported. Error bars represent standard error.