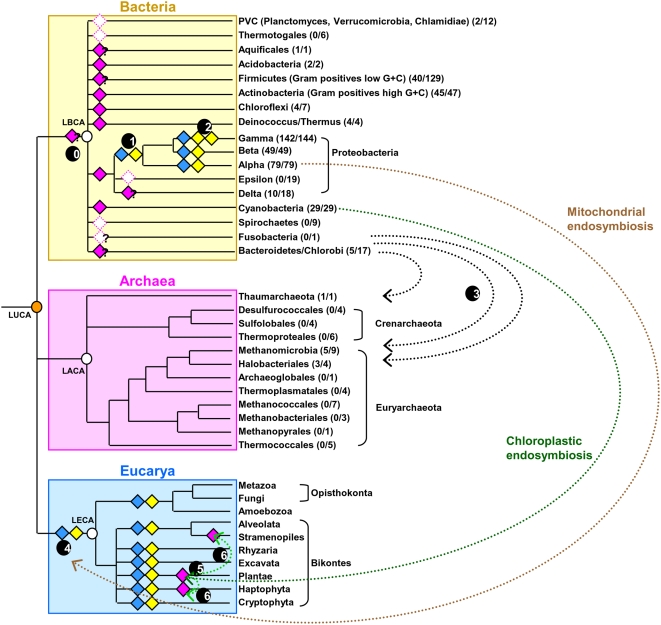
Figure 1. Model for ATC protein evolution.
Schematic representation of the universal tree of life, for which complete genome sequences are available. LUCA (Last Universal Common Ancestor), LECA (Last Eukaryotic Common Ancestor), LACA (Last Archaeal Common Ancestor) and LBCA (Last Bacterial Common Ancestor) are indicated by orange and white circles. For each prokaryotic phylum, the number of genomes encoding at least one ATC homolog with respect to the number of complete available genomes is given between brackets. Filled-diamonds indicate the presence of an ATC encoding gene in the ancestor of a given lineage: pink diamonds designate ATCs; blue diamonds symbolize ATC-I and yellow diamonds represent ATC-II. Dotted empty diamonds symbolize the loss of the corresponding ATC ancestor encoding gene in the lineage. Arrows schematize horizontal gene transfer events (HGT). The distribution and phylogeny of ATC proteins suggest that they originated in the bacterial domain (black circle number 0) and were thus absent in LACA. The inference of ATC encoding genes in the ancestor of most bacterial phyla (i.e. Acidobacteria, Chloroflexi, Actinobacteria, Cyanobacteria, Deinococcus/Thermus, Alpha-, Beta and Gamma-Proteobacteria) suggests that an ATC encoding gene was present in LBCA. Accordingly, the absence of any ATC encoding gene in PVC, Thermotogae, Epsilon-Proteobacteria, and Spirochaetes suggests ancestral losses whereas the ancestral presence or absence of ATC encoding genes in Bacteroidetes/Chlorobi, Delta-Proteobacteria, Aquificae, Fusobacteria and Firmicutes cannot be definitively inferred. In nearly all Alpha-, Beta- and Gamma-Proteobacteria, at least two ATC encoding genes are present, suggesting their presence in the ancestor of these lineages (blue and yellow filled-diamonds). The evolutionary event at the origin of these two copies cannot be definitively inferred (acquisition through HGT of a non-proteobacterial bacterial sequence or duplication of the native copy, black circle number 1). The acquisition of a third copy in Gamma-Proteobacteria through a duplication event or an HGT occurred later (black circle number 2). The presence of few ATC encoding genes in Archaea likely results from several independent HGTs from different bacterial donors (black arrows and black circle number 3). The two ATC encoding genes found in nearly all eukaryotes are orthologs to the two copies found in Alpha-Proteobacteria and were very likely acquired through the mitochondrial endosymbiosis by their last common ancestor (brown arrow, black circle number 4). The third ATC encoding gene found in Plantae was likely acquired through the primary chloroplastic endosymbiosis (dark green arrow and black circle number 5) and spread to other photosynthetic eukaryotes through secondary chloroplastic endosymbioses (black circles number 6).