Abstract
Several l-aminoacyl-tRNA synthetases can transfer a d-amino acid onto their cognate tRNA(s). This harmful reaction is counteracted by the enzyme d-aminoacyl-tRNA deacylase. Two distinct deacylases were already identified in bacteria (DTD1) and in archaea (DTD2), respectively. Evidence was given that DTD1 homologs also exist in nearly all eukaryotes, whereas DTD2 homologs occur in plants. On the other hand, several bacteria, including most cyanobacteria, lack genes encoding a DTD1 homolog. Here we show that Synechocystis sp. PCC6803 produces a third type of deacylase (DTD3). Inactivation of the corresponding gene (dtd3) renders the growth of Synechocystis sp. hypersensitive to the presence of d-tyrosine. Based on the available genomes, DTD3-like proteins are predicted to occur in all cyanobacteria. Moreover, one or several dtd3-like genes can be recognized in all cellular types, arguing in favor of the nearubiquity of an enzymatic function involved in the defense of translational systems against invasion by d-amino acids.
Although they are detected in various living organisms (reviewed in Ref. 1), d-amino acids are thought not to be incorporated into proteins, because of the stereospecificity of aminoacyl-tRNA synthetases and of the translational machinery, including EF-Tu and the ribosome (2). However, the discrimination between l- and d-amino acids by aminoacyl-tRNA synthetases is not equal to 100%. Significant d-aminoacylation of their cognate tRNAs by Escherichia coli tyrosyl-, tryptophanyl-, aspartyl-, lysyl-, and histidyl-tRNA synthetases has been characterized in vitro (3–9). Recently, using a bacterium, transfer of d-tyrosine onto tRNATyr was shown to occur in vivo (10).
With such misacylation reactions, the resulting d-aminoacyl-tRNAs form a pool of metabolically inactive molecules, at best. At worst, d-aminoacylated tRNAs infiltrate the protein synthesis machinery. Although the latter harmful possibility has not yet been firmly established, several cells were shown to possess a d-tyrosyl-tRNA deacylase, or DTD, that should help them counteract the accumulation of d-aminoacyl-tRNAs. This enzyme shows a broad specificity, being able to remove various d-aminoacyl moieties from the 3′-end of a tRNA (4–6, 11). Such a function makes the deacylase a member of the family of enzymes capable of editing in trans mis-aminoacylated tRNAs. This family includes several homologs of aminoacyl-tRNA synthetase editing domains (12), as well as peptidyl-tRNA hydrolase (13, 14).
Two distinct deacylases have already been discovered. The first one, called DTD1, is predicted to occur in most bacteria and eukaryotes (see Table 1). Inactivation of the gene of this deacylase in E. coli (dtd) or in Saccharomyces cerevisiae (DTD1) exacerbates cell growth inhibition by several d-amino acids, including d-tyrosine (6). In fact, in an E. coli Δdtd strain grown in the presence of 2.4 mm d-tyrosine, as much as 40% of the cellular tRNATyr pool becomes esterified with d-tyrosine (10).
TABLE 1.
Distribution of DTD1 and DTD2 homologs in various phylogenetic groups
Homologs of DTD1 and DTD2 were searched for using a genomic Blast analysis against complete genomes in the NCBI Database (www.ncbi.nlm.nih.gov). Values in the table are number of species. For instance, E. coli is counted only once in γ-proteobacteria despite the fact that several E. coli strains have been sequenced.
| DTD1 | DTD2 | DTD1 + DTD2 | None | |
|---|---|---|---|---|
| Bacteria | ||||
| Acidobacteria | 2 | 0 | 0 | 0 |
| Actinobacteria | 27 | 0 | 0 | 8 |
| Aquificae | 1 | 0 | 0 | 0 |
| Bacteroidetes/Chlorobi | 12 | 0 | 0 | 5 |
| Chlamydiae | 1 | 0 | 0 | 6 |
| Chloroflexi | 4 | 0 | 0 | 0 |
| Cyanobacteria | 5 | 0 | 0 | 16 |
| Deinococcus/Thermus | 4 | 0 | 0 | 0 |
| Firmicutes | ||||
| Bacillales | 19 | 0 | 0 | 0 |
| Clostridia | 19 | 0 | 0 | 0 |
| Lactobacillales | 23 | 0 | 0 | 0 |
| Mollicutes | 0 | 0 | 0 | 15 |
| Fusobacteria/Planctomycetes | 2 | 0 | 0 | 0 |
| Proteobacteria | ||||
| α | 6 | 0 | 0 | 55 |
| β | 24 | 0 | 0 | 11 |
| γ | 80 | 0 | 0 | 8 |
| δ | 15 | 0 | 0 | 0 |
| ε | 1 | 0 | 0 | 12 |
| Spirochaetes | 0 | 0 | 0 | 7 |
| Thermotogae | 5 | 0 | 0 | 0 |
| Archaea | ||||
| Crenarchaeota | 0 | 13 | 0 | 0 |
| Euryarchaeota | 1 | 26 | 0 | 2 |
| Nanoarchaeota | 0 | 0 | 0 | 1 |
| Eukaryota | ||||
| Dictyosteliida | 1 | 0 | 0 | 0 |
| Fungi/Metazoa | ||||
| Fungi | 13 | 0 | 0 | 1 |
| Metazoa | 19 | 0 | 0 | 0 |
| Kinetoplastida | 3 | 0 | 0 | 0 |
| Viridiplantae | 4 | 4 | 4 | 0 |
Homologs of dtd/DTD1 are not found in the available archaeal genomes except that of Methanosphaera stadtmanae. A search for deacylase activity in Sulfolobus solfataricus and Pyrococcus abyssi led to the detection of another enzyme (DTD2), completely different from the DTD1 protein (15). Importing dtd2 into E. coli functionally compensates for dtd deprivation. As shown in Table 1, the DTD2 protein has homologs in most archaea and in plants (16).
Several cells contain neither dtd nor dtd2 homologs (Table 1). For instance, 28 cyanobacterial genomes out of the 33 available ones lack any deacylase-like gene. We took Synechocystis sp. PCC6803 as a model and examined whether this cell contained deacylase activity. This led us to the discovery and characterization of a third type of d-tyrosyl-tRNA deacylase (DTD3). This protein, encoded by dtd3, behaves as a metalloenzyme. Sensitivity of the growth of Synechocystis to external d-tyrosine is strongly exacerbated by the disruption of dtd3. Moreover, expression of the Synechocystis DTD3 in a Δdtd E. coli strain, from a plasmid, restores the resistance of the bacterium to d-tyrosine. Finally, using the available genomes, we examined the occurrence of DTD3 in the living world. The prevalence of DTD3-like proteins is surprisingly high. It suggests that the defense of protein synthesis against d-amino acids is universal.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions—Strains and plasmids used in this study are listed in supplemental Table 1. Synechocystis strains were cultivated under shaking at 30 °C with a photon flux density of 50 μmol·m–2·s–1 in BG11 medium (Sigma) modified by the addition of 10 mm NaHCO3, 5 mm Hepes (pH 8), 46 μm H3BO3, 9.1 μm MnCl2, 0.77 μm ZnSO4, 1.9 μm MoNa2O4, 0.32 μm CuSO4, and 0.33 μm Co(NO3)2. For growth on plates, 1.9% agar was added. E. coli strains were cultivated in 2× TY-rich medium or in M9-glucose minimal medium at 37 °C, with shaking.
Preparation of Crude Extracts—d-Tyr-tRNATyr deacylase activity in a fresh crude extract of Synechocystis strain PCC6803 was assayed using d-[3H]Tyr-tRNATyr as substrate. A culture of cyanobacteria (100 ml) was centrifuged at 9800 × g for 15 min at 4 °C, and the cell pellet was resuspended in 20 mm Tris-HCl (pH 6.8) containing or not 160 μm NiCl2. The same protocol was applied for comparison of the deacylase activity in extracts of strains PCC6803 and PCC6803ΔDTD3 except that 40 μm CoCl2 was added to the buffer instead of 160 μm NiCl2. Final volumes of the bacterial suspensions were adjusted to obtain an optical density (OD) of ∼100 at 750 nm. Cells were disrupted by sonication (3 min, 0 °C), and debris was removed by centrifugation (10 min, 21,400 × g, 4 °C).
To obtain crude extracts of E. coli cells transformed with pET3alpa, pET3alpa::1786, or pET3alpa::0208, bacteria were grown at 37 °C in 50 ml of 2× TY medium containing 100 μg/ml ampicillin. When the stationary phase of growth was reached, isopropyl 1-thio-β-d-galactopyranoside (IPTG)2 was added at a final concentration of 1 mm, and the culture was further incubated at 4 °C for 5 h with shaking. After centrifugation at 9800 × g for 10 min at 4 °C, bacteria were resuspended in 20 mm Tris-HCl (pH 6.8) containing 0.1 mm phenylmethylsulfonyl fluoride (PMSF) and disrupted following the same protocol as that used with the Synechocystis cells. Total amounts of proteins in all extracts were measured by using the Bradford protein assay (Bio-Rad), with bovine serum albumin as standard.
Measurement of Deacylation Rates—d-[3H]Tyr-tRNATyr (500 Ci/mol), l-[14C]Tyr-tRNATyr (434 Ci/mol), and diacetyl-l-[14C]Lys-tRNALys (310 Ci/mol) were prepared from E. coli tRNAs as described previously (6, 11, 14, 16).
To assay deacylase activity during the purification of the DTD3 protein from Synechocystis extracts, protein samples were diluted in 20 mm Tris-HCl (pH 6.8) containing 160 μm NiCl2 (TN buffer). This solution was further diluted 4-fold in a 100-μl assay. Finally, d-Tyr-tRNATyr hydrolysis was followed for 5 min at 28 °C in 20 mm Tris-HCl (pH 7.8) containing 40 μm NiCl2 and 100 nm d-[3H]Tyr-tRNATyr. During the purification of DTD3 from recombinant E. coli cells, d-Tyr-tRNATyr hydrolysis was followed as described above, except that NiCl2 was omitted from the assay mixture and from the enzyme dilution buffer.
Unless otherwise stated, the activity of the purified DTD3 protein was measured under initial conditions for 5 min, at 28 °C, in 100-μl assays containing 100 nm substrate, 20 mm Tris-HCl (pH 7.8), 3 mm MgCl2, and 40 μm CoCl2. Before use, the enzyme was diluted in 20 mm Tris-HCl (pH 6.8) containing 200 μg/ml bovine serum albumin and 40 μm CoCl2.
In all cases, the deacylation reaction was quenched by the addition of 340 μl of ethanol, 14 μl of sodium acetate 3 m (pH 4.8), and 20 μl of carrier RNA (4 mg/ml) from yeast (Roch-Light Ltd.). Samples were centrifuged (20 min, 21,400 × g, 4 °C), and the radioactivity in the supernatant was measured by scintillation counting (11). One unit corresponds to the enzyme activity capable of hydrolyzing 1 pmol of d-Tyr-tRNATyr per min in the above conditions. For each rate measurement, two independent experiments were performed. Mean values are given with associated ranges.
To measure the catalytic constants of purified deacylase in the reaction of d-Tyr-tRNATyr hydrolysis, initial rates were followed as above, with cobalt, in miniaturized (20 μl) assays containing increasing concentrations of substrate (90–6000 nm) and a constant deacylase concentration of 165 pm. Km and kcat values with standard deviations were derived from iterative nonlinear fits of the theoretical Michaelis equation to the experimental values using the Levenberg-Marquardt algorithm (17).
Purification of d-Aminoacyl-tRNA Deacylase from Synechocystis—The culture (18 liters) of Synechocystis was centrifuged (11,300 × g, 10 min, 4 °C). The cell pellet (∼18 g of wet weight) was resuspended in 180 ml of TN buffer containing 0.1 mm PMSF. Cells were disrupted by sonication (10 min, 0 °C), and the extract was centrifuged (9800 × g, 20 min, 4 °C). Nucleic acids were precipitated by the addition of streptomycin (30 mg/ml) to the supernatant. Then the sample was centrifuged for 20 min at 9800 × g (4 °C). The supernatant was diluted to 400 ml and applied on a Q-Sepharose FastFlow column (GE Healthcare) of 450 ml equilibrated in TN buffer. Elution was carried out at a flow rate of 2 ml/min with a 6-liter linear gradient of 0–335 mm NaCl in the same buffer. d-Tyr-tRNATyr deacylase activity was recovered between 200 and 250 mm NaCl. Fractions containing activity were pooled (510 ml) and dialyzed twice against 5 liters of TN buffer. To concentrate the sample, the pooled fractions were applied on a 10-ml Q-Sepharose FastFlow column equilibrated in TN buffer. Elution was carried out at a flow rate of 1 ml/min with a 200-ml linear gradient of 0–335 mm NaCl in the TN buffer. Active fractions were pooled (20 ml). The resulting sample was concentrated to a volume of ∼5 ml with an Amicon Ultra-4 filter (cut-off 10 kDa; Millipore) and loaded onto a Superdex 75 column (1.6 × 60 cm; GE Healthcare) equilibrated in TN buffer containing 150 mm NaCl. The column was eluted at a flow rate of 0.4 ml/min. Fractions with deacylase activity were pooled (11 ml), and the sample was concentrated to 1.5 ml with Amicon Ultra-4 filters. Ammonium sulfate was added to the resulting sample at a final concentration of 1.7 m. After centrifugation of the insoluble material (20 min, 21,400 × g, 4 °C), the supernatant was applied to a Bakerbond Wide-Pore Hi-propyl C3 (Baker) column equilibrated in TN buffer containing 3 m ammonium sulfate. Elution was carried out at a flow rate of 0.1 ml/min with a 20-ml decreasing linear gradient of ammonium sulfate (from 3 to 0 m) in TN buffer. Fractions containing deacylase activity were precipitated in 10% trichloroacetic acid and 0.01% Triton X-100 and submitted to SDS-PAGE.
DNA Extraction—A culture of cyanobacteria (6 ml) was centrifuged (9200 × g, 5 min, 4 °C). The cell pellet was resuspended in 300 μl of 50 mm Tris-HCl (pH 7.5) containing 20% saccharose, 5 mm EDTA and 5 mg/ml lysozyme. After 45 min of incubation of the mixture at 37 °C, 40 μl of 10% SDS and 300 μl of a solution containing 150 mm NaCl and 10 mm EDTA were added. Then two phenol extractions were performed. To remove phenol, 600 μl of diethyl ether were added and vigorously mixed with the sample. After centrifugation (21,400 × g, 5 min, 20 °C), the aqueous phase was precipitated with 500 μl propan-2-ol and centrifuged once more. The DNA pellet was washed with 70% ethanol, dried, and resuspended in 50 μl of 20 μg/ml RNase A (Roche Applied Science). After a 20-min incubation at 37 °C, the sample was stored at 4 °C.
Cloning of the sll1786 and sll0208 Genes—The genomic DNA of strain PCC6803 was used as a template to amplify the sll1786 and sll0208 genes. The primers chosen for PCR amplification were sll1786_NdeI_for plus sll1786_NotI_rev for the insertion of sll1786 into the pET3alpa plasmid, sll1786_XbaI_for plus sll1786_NotI_rev for the insertion of sll1786 into pET15blpa, and sll0208_NdeI_for plus sll0208_NotI_rev for the insertion of sll0208 into pET3alpa (see supplemental Table 2 for the sequences of the primers). The resulting amplified fragments were purified by using the Qiagen PCR purification kit 50 and digested by either NdeI plus NotI or by XbaI plus NotI. The two digestion products were inserted into plasmids pET3alpa and pET15blpa to give plasmids pET3alpa::1786, pET15blpa::1786, and pET3alpa::0208. With each plasmid, the integrity of the cloned gene was verified by DNA sequencing.
Purification of the sll1786 Gene Product—E. coli strain K37ΔRecAΔTyrHλDE3 (15) was transformed by plasmid pET3alpa::1786. Transformed cells were grown at 37 °C until the stationary phase in 2 liters of 2× TY medium containing 100 μg/ml ampicillin. Then 1 mm of IPTG was added, and the culture was left to incubate for 5 h at 4 °C. Cells were harvested by centrifugation for 10 min at 9800 × g (4 °C). The cell pellet was resuspended in 20 mm Tris-HCl (pH 6.8) containing 0.1 mm PMSF to obtain an OD of ∼100 at 650 nm. Cells were disrupted by sonication (10 min, 0 °C), and debris was removed by centrifugation (20,400 × g, 10 min, 4 °C). Nucleic acids were precipitated by addition of streptomycin (30 mg/ml) to the supernatant, which was then centrifuged for 20 min at 20,400 × g (4 °C). The resulting supernatant was applied on a Q-Sepharose FastFlow column (2.5 × 10 cm) equilibrated in 20 mm Tris-HCl (pH 6.8). Elution was performed at a flow rate of 1 ml/min with a 1-liter linear gradient of 0–0.5 m NaCl. Enzyme activity was recovered between 200 and 250 mm NaCl. Active fractions were pooled and directly applied on a hydroxylapatite column (1.1 × 2 cm; Bio-Rad) equilibrated in 20 mm Tris-HCl (pH 6.8). This column was eluted at a flow rate of 0.2 ml/min with 20 ml of a linear sodium phosphate buffer gradient (0–0.5 m sodium phosphate (pH 6.4), in 20 mm Tris-HCl). Fractions with enzyme activity recovered between 250 and 275 mm phosphate were pooled. According to SDS-PAGE analysis, the purified protein was at least 90% homogeneous. The purification procedure yielded 30 mg of protein. Finally, the protein sample was concentrated to 15 ml with an Amicon Ultra-4 filter, dialyzed against 20 mm Tris-HCl (pH 6.8) containing 60% glycerol, and stored at –20 °C.
Disruption of the sll1786 Gene—The 800 bp downstream from the sll1786 ORF were amplified by PCR using genomic DNA of Synechocystis as template and oligonucleotides Av_ SacII and Av_NotI as primers. The resulting fragment was digested by SacII plus NotI and introduced into the corresponding sites on pET3alpa to produce pET3alpa::Av. The insert was verified by DNA sequencing. The 800 bp upstream of sll1786 were amplified with the primers Am_NdeI and Am_ KpnI. The PCR product was digested by NdeI plus KpnI and cloned into the corresponding sites on pET3alpa::Av, thus creating pET3alpa::Am::Av. The integrity of the insert was verified by DNA sequencing. The kanamycin resistance sequence of pUC4K (18) was amplified with the primers Kan_KpnI and Kan_SacII. The resulting DNA fragment was digested by KpnI and SacII. Then the fragment was ligated with pET3alpa::Am::Av previously digested by the same enzymes to produce pET3alpa::Am::Kan::Av. After transformation of E. coli with the ligation mixture, the clones containing plasmid pET3alpa::Am::Kan::Av were selected on LB agar containing 25 μg/ml kanamycin.
An exponentially growing Synechocystis sp. PCC6803 culture (40 ml, OD750 ∼0.2) was centrifuged (1400 × g, 20 °C). After resuspension in 5 ml of fresh medium, the cells were exposed to 50 μg of pET3alpa::Am::Kan::Av DNA. The transformation mixture was incubated for 6 h at 30 °C under light and shaking. 100 μl of the transformation mixture was then plated on a membrane filter (ME25, Whatman) and laid on a Petri dish containing modified BG11 agar medium. The dish was incubated at 30 °C during 60 h under light, and the membrane filter was shifted on modified BG11 agar plate supplemented with 30 μg/ml kanamycin. After 9 days of incubation at 30 °C with light, mutants were streaked on BG11 agar medium containing 30 μg/ml kanamycin. The replacement of sll1786 by the kanamycin resistance gene was verified by PCR using oligonucleotides kan-2 plus 30s_6 and 1786_1 plus 1786_5.
Measurement of Generation Times—E. coli generation times were measured as described previously (16). To start the growth of cyanobacteria, Synechocystis strains PCC6803 and PCC6803ΔDTD3 were inoculated in 20 ml of the medium under study. The inoculates were such that an OD750 of 0.2 was reached. The OD of the culture was then measured every day for 7 days. Generation times with standard deviations were deduced from fits of these measurements to an exponential function (17).
Mass Spectrometry—Mass spectrometry-based protein identification was done with the standard protein mass fingerprint method. Briefly, gel bands were cut and subjected to reduction (10 mm dithiothreitol, 56 °C, 30 min) followed by alkylation (55 mm iodoacetamide, room temperature, 20 min) and digestion by trypsin (12.5 ng/μl, 37 °C, 2 h; Roche Applied Science). After extraction (1% formic acid (FA) in water, followed by 1% FA in acetonitrile), the resulting peptide mixture was lyophilized, dissolved in 1% FA water, and desalted with a C18 micropipette tip (Agilent) using 50/50/1 water/acetonitrile/FA as the elution solvent. The masses of the peptides were then measured on a Q-TOF Premier (Waters) mass spectrometer. The mass list was submitted to a data base search using the Mascot search engine against the NCBInr data base. The search took into account two possible missed cleavages, carbamidomethylation of cysteines and optional oxidation of methionines.
To measure the mass of the native protein, the protein-metal cations complexes were desalted using Micro-BioSpin 6 chromatography columns (BioSpin). First, a concentrated stock of ammonium acetate (1 m) was chromatographed through a Chelex 100 column (0.2 × 7 cm; Bio-Rad) to remove contaminating metal ions. The recovered buffer was diluted 100-fold with purified water. This solution was used to equilibrate BioSpin columns, as indicated by the manufacturer for buffer exchange (four equilibrations with 500 μl of 10 mm ammonium acetate). Then 70 μl of the initial protein solution were loaded onto a first chromatography column and eluted by centrifugation (2 min, 1000 × g). This step was repeated on a new chromatography column. When necessary, denaturation of the protein to remove noncovalently bound metal ions was done by adding to the protein solution an equal volume of 4% FA in acetonitrile. The solutions were analyzed on two mass spectrometers. A Q-TOF Premier (Waters) mass spectrometer was used for the quantitation of the number of metal ions present on the protein (19). The mass spectra were recorded without any selection in the first quadrupole, without collision energy and with a mass range of m/z 1000–4000. An FT-ICR Apex III (Bruker) mass spectrometer equipped with a 7.0-tesla actively shielded magnet was used for high resolution experiments to identify the metal ions bound to the protein. The spectra were recorded with a mass range of m/z 1000–6000. For both instruments, the solution was loaded in a nano-electrospray needle and set in the source of the mass spectrometer. Source temperature and voltages were adjusted to minimize complex dissociation.
Atomic Absorption Spectroscopy—Flame atomic absorption spectroscopy measurements were performed using a Varian AA220 spectrophotometer equipped with an air-acetylene burner. Zinc and cobalt contents were measured in the peak mode, with 100-μl injections at 213.9 and 240.7 nm, respectively. Metal concentrations were deduced from two independent experiments in which each measurement was made in triplicate. Mean values are given with associated ranges. When enzyme was dialyzed prior to atomic absorption measurements, the dialysis buffer was deprived of contaminating divalent cations as described above for the preparation of the ammonium acetate solution.
RESULTS
Purification of d-Tyr-tRNATyr Deacylase Activity from Synechocystis sp. PCC6803 Crude Extracts—Strain Synechocystis sp. PCC6803 was cultured in modified BG11 medium (see “Experimental Procedures”). Cells were harvested after 1 week of growth, and isolation of the enzyme supporting the deacylase activity was attempted at 4 °C.
We observed that purification of the protein could not be achieved because of spontaneous loss of the activity in the cell extract. To possibly protect deacylase activity, several compounds were added to the extract, including metals, EDTA, and reducing agents. The addition of Ni2+, Mn2+ or Co2+, but not Zn2+ protected the activity enough to trace the enzyme over weeks. As will be discussed below, this behavior reflects an interference of transition metal ions with the activity of the deacylase. Eventually, with the presence of NiCl2 in all purification buffers and in the activity assay, isolation of a deacylase sample could be carried out within 3 weeks, as follows.
d-Tyr-tRNATyr deacylase activity in a fresh crude extract was assayed using d-[3H]Tyr-tRNATyr as substrate. Deacylation of the d-aminoacyl-tRNA occurred at a rate of 240 units per mg of protein (28 °C, 20 mm Tris-HCl (pH 7.8), 40 μm NiCl2). At the end of the deacylation reaction, it was verified that the tRNA could still be fully aminoacylated in the presence of E. coli tyrosyl-tRNA synthetase and l-[14C]tyrosine.
To isolate further this activity, a crude extract was prepared from an 18-liter culture. Nucleic acids were precipitated by streptomycin. After chromatographies on two successive Q-Sepharose columns, one Superdex 75 and one Hi-propyl column, the specific deacylase activity was enriched by nearly 150-fold, compared with the crude extract. Active fractions recovered from the last column were analyzed by SDS-PAGE. Several protein bands were visible. The intensities of two of them, with migrations around 30 kDa, varied proportionally with the deacylase activity along the elution profile of the column. The corresponding proteins were recovered from the gel and submitted to tryptic digestion. Mass spectrometry analysis of the resulting peptides identified the sll0208 and sll1786 ORFs from Synechocystis sp. PCC6803 as the sources of the two proteins.
The two ORFs were amplified by PCR and inserted into the expression vector pET3alpa. E. coli strain K37ΔRecAΔTyrHλDE3 (Δdtd) was transformed with the resulting plasmids (pET3alpa::0208 or pET3alpa::1786). Upon induction with IPTG, each strain overexpressed a soluble protein having the expected mass of ∼30 kDa. d-Tyrosyl-tRNATyr deacylase activity was assayed in crude extracts, in the presence of 40 μm NiCl2. In the crude extract of K37ΔRecAΔTyrHλDE3 containing pET3alpa:1786, the specific deacylase activity (28 °C, 20 mm Tris-HCl (pH 6.8)) was at least 1500-fold higher (138,000 units/mg protein) than those measured using the strains transformed by either pET3alpa:0208 or the control plasmid pET3alpa (<90 units/mg). Thus, we concluded that the sll1786 ORF, not the sll0208 one, accounted for d-tyrosyl-tRNA deacylase activity in the cyanobacterium. The product of this gene, with a theoretical molecular mass of 29,258 Da, was named DTD3.
Effect of Metal Ions on the Activity of DTD3—The DTD3 protein was purified from E. coli cells K37ΔRecAΔTyrHλDE3 harboring pET3alpa::1786. Contrary to the purification of the protein from the Synechocystis extract, we did not need to add a metal to the E. coli extract to pursue the purification procedure. The main reason is that purification from the E. coli extract required a much shorter time (3 days) than that necessary when starting from a Synechocystis extract. In an assay mixture without any added divalent metal, the specific activity of the nearly homogeneous native enzyme was 0.024 ± 0.003 s–1. In the presence of 3 mm MgCl2, a counterion of the polyanionic d-aminoacyl-tRNA substrate, this value increased to 0.034 ± 0.004 s–1.
High resolution mass spectrometry of the purified protein under native conditions showed an isotopically resolved isotope distribution centered at 29,320.5 ± 0.5 Da. In denaturing conditions, the same sample led to an isotopic distribution centered at 29,258.5 ± 0.5 Da, very close to the theoretical molecular mass (29,258.0 Da). Isotopic fitting of this experimental distribution with the theoretical distribution expected for DTD3 agrees within 0.2 Da. A mass difference of 62.0 Da is observed between the native and denatured protein. Because the expected mass difference for the addition of 1 Zn2+ ion is 63.4 Da, this could indicate the presence of 1 Zn2+ ion per polypeptide chain. To determine whether a zinc ion was indeed associated with the native protein, flame atomic absorption spectroscopy was used. A stoichiometry of 0.9 ± 0.2 mol of zinc/mol of protein was found.
The effects of various compounds (Co2+, Cu2+, Fe2+, Mn2+, Ni2+, Zn2+, or EDTA) on the enzyme activity were investigated. The enzyme was incubated for 10 min with 160 μm of each compound, and its activity was measured for 5 min after a 4-fold dilution in the assay mixture (20 mm Tris-HCl (pH 7.8), 3 mm MgCl2). With Co2+, Mn2+, and Ni2+, the activity rate value reached 0.85, 0.95, and 0.70 s–1, respectively. With Cu2+,Fe2+, Zn2+, or EDTA, activity values (0.03, 0.11, 0.007, and 0.005 s–1, respectively) remained close to that of the native enzyme (0.034 s–1). The above experiments indicated that the transition metal ions Co2+, Mn2+, and Ni2+ markedly contributed to the expression of the deacylase activity.
To determine the minimal concentrations of Co2+, Mn2+, and Ni2+ needed to stimulate the activity of DTD3, various concentrations of these ions were added to the enzyme. In this set of experiments, metal concentrations were the same in the assay and in the buffer used to dilute the enzyme solution prior to the assay. In all cases, the initial activity (0.034 s–1) was increased by a factor of around 25 at saturation of the metal. The dose-response curves were fitted to a hyperbolic function (17). Half-maximal responses were at metal concentrations of 2.8 ± 0.6, 4.9 ± 0.8, and 25 ± 5 μm, for Co2+, Mn2+, and Ni2+, respectively.
Because the purified native protein contained ∼1 zinc ion per molecule, we wondered whether the stimulating effect obtained upon cobalt, manganese, or nickel addition was caused by substitution of this protein-bound zinc or by occupation of additional metal sites. Cobalt was chosen as an example. A 50 μm enzyme sample in 500 μl was dialyzed against 12 ml of a 20 mm Tris-HCl buffer (pH 6.8) containing 3 mm MgCl2 and 20 μm CoCl2. The buffer contained less than 0.5 μm of contaminating zinc. After overnight dialysis at 4 °C, cobalt and zinc amounts were determined by flame atomic absorption spectroscopy. Cobalt concentrations outside and inside the dialysis bag were 18 ± 1 and 72 ± 2 μm, respectively. These values indicated 1.1 ± 0.1 bound cobalt per polypeptide chain. Zinc concentration inside the bag was 45 ± 5 μm, corresponding to 0.9 ± 0.1 zinc per polypeptide chain. We therefore concluded that the zinc ion in the native deacylase could not be spontaneously displaced from the protein by cobalt and that the cobalt occupied a second metal site.
As reported above, in the presence of 40 μm zinc in the assay, the value of the enzyme activity was near 10–2 s–1. To study further this inhibitory effect of zinc, the DTD3 enzyme (55 μm in 700 μl) was dialyzed against 60 ml of a 20 mm Tris-HCl buffer (pH 6.8) containing 3 mm MgCl2 and 5 μm ZnCl2. At the end of the dialysis, free zinc was 4.3 ± 0.2 μm, and enzyme-bound zinc amounted to 1.9 ± 0.2 mol per mol of enzyme, as deduced from atomic absorption spectroscopy measurements. The dialyzed enzyme was diluted 150,000-fold in the presence of 160 μm of either CoCl2, MnCl2, NiCl2, or ZnCl2 and immediately assayed for 5 min in the presence of 40 μm of these metals. In all cases, the activity remained low (<0.01 s–1). All these data indicate that DTD3 can bind two zinc ions at the same time, that the zinc2-protein complex is poorly active, and that the second zinc cannot be easily exchanged with cobalt, manganese, or nickel.
Another possibility is that upon the binding of 2 zinc ions, the enzyme irreversibly looses activity. To determine whether the inhibition of DTD3 by zinc was reversible, an enzyme sample loaded with 2 zincs per polypeptide chain was incubated in the presence of 1 mm EDTA. Native mass spectrometry analysis of the enzyme treated with EDTA for 7 days showed a single peak at 29,255 ± 3 Da, very close to the theoretical value for the apoenzyme. Thus, the week-long EDTA treatment appeared to have removed any metal from the enzyme. The EDTA-treated enzyme reached an activity of 1.5 ± 0.4 s–1 when assayed in the presence of 40 μm CoCl2. Control experiments with 40 μm of either EDTA or ZnCl2 in the assay displayed low enzyme activity values (<0.005 s–1). In a final experiment, the EDTA-treated DTD3 was submitted to size-exclusion chromatography on a Micro-BioSpin 6 column equilibrated in 20 mm Tris-HCl (pH 6.8) containing 40 μm CoCl2. According to mass spectrometry, the recovered protein was loaded with 2 Co2+ ions (29,371 ± 3 Da). Altogether, these results indicate the following: (i) the two-zinc enzyme was not irreversibly inactivated; (ii) both metals could be removed with EDTA and substituted with cobalt; (iii) the metal-free apoenzyme is poorly active; and (iv) the two-cobalt enzyme regains an activity value similar to that of the enzyme carrying both 1 zinc and 1 cobalt.
At this stage, we may imagine that the loss of deacylase activity in the Synechocystis extracts was caused by a slow poisoning of the enzyme by contaminating zinc at the second metal site of the enzyme. The addition of nickel, cobalt, or manganese to the extracts appeared sufficient to compete with the Zn2+ ions for access to this second metal center and to protect the activity of the enzyme against inhibition.
Specificity and Catalytic Constants of DTD3—The catalytic constants of native DTD3 were measured in the presence of 40 μm CoCl2 (28 °C, 20 mm Tris-HCl (pH 7.8), 3 mm MgCl2). By varying the d-Tyr-tRNATyr concentration (from 90 to 6000 nm) in the presence of 165 pm purified enzyme, Km and kcat values of 0.8 ± 0.2 μm and 7.5 ± 0.7 s–1 were obtained. These values are close to those measured with E. coli DTD1 (11). Rates of d- and l-Tyr-tRNATyr hydrolysis by DTD3 were compared in a 5-min assay, including 20 mm Tris-HCl (pH 6.8), 3 mm MgCl2,40 μm CoCl2, 100 nm substrate, and 0.2 to 40 nm of enzyme (Table 2). The initial rate of enzymatic d-Tyr-tRNATyr deacylation was at least 95-fold faster than that of l-Tyr-tRNATyr deacylation. Under the same assay conditions, diacetyl-l-Lys-tRNALys resisted the action of the enzyme, with a deacylation rate at least 900-fold lower than that measured with d-Tyr-tRNATyr as substrate. Finally, we verified that after deacylation by homogeneous DTD3, the tRNATyr product was intact, as assessed by its capacity to be aminoacylated by E. coli tyrosyl-tRNA synthetase.
TABLE 2.
Substrate specificity of DTD3
Initial rates of hydrolysis by purified Synechocystis DTD3 protein (0.2–40 nm) were measured at 28 °C for 5 min in the presence of 20 mm Tris-HCl (pH 7.8), 100 nm of the substrate under study, 3 mm MgCl2, 40 μm CoCl2, and 50 μg/ml bovine serum albumin. Values shown are the mean of two independent experiments. Ranges are within 15%.
| Substrate | Initial rates |
|---|---|
| s-1 | |
| d-Tyr-tRNATyr | 0.81 |
| l-Tyr-tRNATyr | 8.4 × 10-3 |
| Diacetyl-l-Lys-tRNALys | <0.9 × 10-3 |
Inactivation of the Chromosomal dtd3 Gene—The dtd3 gene of Synechocystis sp. PCC6803 was deleted and replaced by a kanamycin resistance cassette. In a crude extract of the resulting strain PCC6803ΔDTD3, the d-Tyr-tRNATyr deacylase activity (1.74 ± 0.24 units/mg) was nearly 200-fold smaller than that in a crude extract of the parental strain (318 ± 24 units/mg). Measurements were made in the presence of 40 μm CoCl2 in the assay. Thus, we concluded that, in Synechocystis sp. PCC6803, the dtd3 gene is responsible for almost all the d-Tyr-tRNATyr deacylase activity.
The generation times of strains PCC6803 and PCC6803ΔDTD3 were compared in different culture media (Table 3). They were identical (27 ± 3 h) in modified BG11 medium (see “Experimental Procedures”). In contrast, upon addition to the medium of 5 μm d-tyrosine only, the growth of PCC6803ΔDTD3 cells was strongly impaired (49 ± 3 h) as compared with that of the parent strain (29 ± 1 h). At 80 μm d-tyrosine, the Δdtd3 cells stopped growing. On the other hand, with 80 μm l-tyrosine, there was no effect on the growth. These experiments showed that, in vivo, DTD3 protects the cyanobacterium against the toxicity of d-tyrosine.
TABLE 3.
Effect of d- or l-tyrosine on the growth of various Synechocystis strains
Cells were grown at 30 °C in modified BG11 medium, under light, in the presence or absence of d- or l-tyrosine. Cells were pre-grown in the growth medium under study. Inoculations were adjusted to an OD750 of 0.2. Samples were withdrawn from the culture every day during 7 days, and generation times were deduced from OD750 measurements.
|
Generation time
|
||
|---|---|---|
| PCC6803 | PCC6803ΔDTD3 | |
| h | h | |
| No tyrosine | 26 ± 2 | 27 ± 2 |
| 80 μml-Tyr | 26 ± 1 | 25 ± 1 |
| 5 μmd-Tyr | 29 ± 1 | 49 ± 3 |
| 10 μmd-Tyr | 27 ± 3 | 70 ± 9 |
| 15 μmd-Tyr | 25 ± 3 | 104 ± 7 |
| 20 μmd-Tyr | 25 ± 3 | 142 ± 9 |
| 30 μmd-Tyr | 28 ± 3 | 211 ± 12 |
| 80 μmd-Tyr | 27 ± 2 | No growth |
Expression of Synechocystis DTD3 Cures E. coli for Inactivation of Its Endogenous Deacylase Gene (dtd)—In minimal medium, the generation time of the E. coli Δdtd strain K37ΔRecAΔTyrHλDE3 is significantly increased (1.5-fold) upon addition of millimolar amounts of d-tyrosine (16). Thus, this strain was used to examine whether introduction of DTD3 could confer E. coli resistance against d-tyrosine.
Strain K37ΔRecAΔTyrHλDE3 was transformed with pET3alpa::1786. Because the resulting strain grew very slowly in minimal medium, we suspected that DTD3 overproduction was toxic for the bacterium. In truth, overproduction of a plant DTD2 homolog (16) as well as overexpression of the endogenous DTD1 enzyme itself (11) were already observed to cause inhibition of the growth of E. coli.
To reduce DTD3 expression to tolerable levels, we introduced the sll1786 ORF into plasmid pET15blpa, which differs from the plasmid pET3alpa by the presence of lacI (20). The generation time of strain K37ΔRecAΔTyrHλDE3 containing pET15blpa::1786 was slightly longer (111 min) than that of the same strain carrying pET15blpa (95 min). Nevertheless, we were able to observe that the E. coli strain harboring the Synechocystis ORF had become fully resistant to 2.4 mm d-tyrosine, whereas the control was sensitive (Table 4). In agreement with these phenotypes, d-Tyr-tRNATyr activity in the E. coli strain overexpressing the dtd3 gene was 2400-fold larger than that in the cell containing the control plasmid pET15blpa (Table 4). These results indicate that dtd3 overexpression rescues an E. coli Δdtd mutant from its sensitivity to d-tyrosine.
TABLE 4.
Effect of d-tyrosine on the growth of various E. coli strains
Cells were grown at 37 °C in M9-glucose minimal medium containing 100 μg/ml ampicillin in the presence or absence of 2.4 mm d-tyrosine. Cells were pre-grown overnight in the culture medium under study. Inoculations were adjusted to an OD650 of 0.05. Samples were withdrawn from the culture every 90 min during 540 min, and generation times were deduced from OD650 measurements. At the end of the experiment (0.8 OD650), to measure deacylase activities in extracts, 200-ml aliquots were withdrawn from the control cultures without d-tyrosine. Bacteria were resuspended in 20 mm Tris-HCl (pH 6.8) containing 40 μm CoCl2 and 0.1 mm PMSF. Cells were disrupted as described under “Experimental Procedures.” Deacylase activity was measured in the presence of 40 μm CoCl2.
|
Strain
|
d-Tyr-tRNATyr deacylase activity
|
Generation time
|
||
|---|---|---|---|---|
| Control | d-Tyrosine | |||
| units/mg protein | min | |||
| K37ΔRecAλDE3 (pET15blpa) | 72 ± 12 | 94 ± 7 | 97 ± 6 | |
| K37ΔRecAΔTyrHλDE3 (pET15blpa) | 21 ± 3 | 95 ± 8 | 141 ± 7 | |
| K37ΔRecAΔTyrHλDE3 (pET15blpa::1786) | 51,000 ± 3000 | 111 ± 7 | 112 ± 5 | |
DISCUSSION
This study establishes that Synechocystis sp. PCC6803 possesses a d-Tyr-tRNATyr deacylase distinct from the other two deacylases already documented, DTD1 and DTD2. There are no significant amino acid sequence homologies between the three types of deacylase. The novel deacylase, DTD3, confers Synechocystis resistance against d-tyrosine. Moreover, its overexpression complements the absence of dtd in the context of E. coli. DTD3, which behaves as a monomer, copurifies with one firmly bound zinc ion. This metal ion can be removed by prolonged exposure to EDTA. Upon this treatment, the activity decreases from 0.025 to less than 0.005 s–1. We observed that native DTD3 possesses a second metal ion-binding site. The enzyme activity strongly depends on the nature of the second metal occupying this secondary site. Although enzyme species containing simultaneously Zn2+ and either Co2+, Ni2+, or Mn2+ exhibit high activity values (∼1 s–1), the zinc2-protein complex is poorly active (0.007 s–1). This behavior distinguishes DTD3 from DTD2. Indeed, DTD2 needs two zincs to express full activity (15).
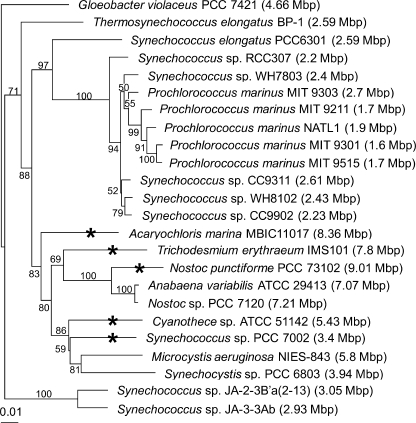
Homologs of Synechocystis dtd3 are recognizable in the 33 available cyanobacterial genomes. On the other hand, 5 of these 33 genomes also display a dtd homolog (Fig. 1). It is possible that dtd was present in the cyanobacterial common ancestor and was then lost in most lineages. In support of this scenario, we have observed that the sequences of the cyanobacterial dtd-like gene products are close to each other, contradicting the idea of late, independent appearances in a limited set of cyanobacteria. However, horizontal gene transfer between cyanobacteria cannot be excluded. Finally, genome sizes of the cyanobacteria possessing both dtd and dtd3 are systematically rather large (Fig. 1). This feature may explain the capacity of these cells to have kept or acquired two redundant deacylase activities.
FIGURE 1.
Distribution of DTD1 homologs in the phylum of cyanobacteria. The 16 S rDNA phylogenetic tree of entirely sequenced cyanobacteria genomes was constructed using the ribosomal data base project web site. For the sake of clarity, several Prochlorococcus and Synechococcus species, whose genomes are highly homologous to those of other species, were omitted from the figure. The concomitant presence of a DTD1 and of a DTD3 homolog is indicated with an asterisk. The genome size of each cyanobacterium is specified in brackets.
The E. coli genome includes three genes homologous to dtd3. The products of these genes, TatD, YjjV, and YcfH, share 35, 29, and 40% sequence identity with Synechocystis DTD3, respectively. The tatD gene is encoded in an operon that also contains the tatA, tatB, and tatC genes, known to encode essential components of the Tat (twin arginine translocation) export pathway. According to Wexler et al. (21), TatD is a cytoplasmic protein, whereas the products of the other genes in the operon are probably membrane proteins. Actually, TatD and the ortholog of TatD in S. cerevisiae have been reported to display DNase activity in vitro in the presence of Mg2+ ions (21, 22). In experiments not shown, we have produced homogeneous TatD, YjjV, and YcfH and assayed their deacylase activity in the presence of either Co2+, Cu2+, Mn2+, Ni2+, or Zn2+. TatD and YjjV were never active. In the case of YcfH, d-tyrosyl-tRNA deacylase activity could be detected provided either cobalt, manganese, or nickel was added. Under standard assay conditions, in the presence of 40 μm CoCl2, an initial rate value of 0.024 s–1 could be estimated. For comparison, a rate value of ∼1 s–1 was measured with Synechocystis DTD3 under similar conditions. After d-Tyr-tRNATyr deacylation under the action of the YcfH protein, the tRNATyr product could still be fully aminoacylated by E. coli tyrosyl-tRNA synthetase. This control experiment excludes the possibility that YcfH acted as an RNase enzyme.
The three-dimensional structures of E. coli TatD, YjjV, and YcfH are available (PDB codes 1xwy, 1zzm, and 1yix, respectively). The three proteins belong to a superfamily of metal-dependent hydrolases (23). A large subset of the members of this superfamily has a binuclear metal center. The three-dimensional folds of TatD, YjjV, and YcfH are very close to each other, with a root mean square deviation of the order of 1.7 Å over more than 250 residues (total length is between 259 and 265 amino acids). Because of the high sequence identity between DTD3 and these proteins, it is likely that DTD3 adopts a similar fold. Therefore, the three-dimensional structure of the DTD3 deacylase would be fully distinct from those of the DTD1 (24) and DTD2 (PDB accession number 1YQE) deacylases. YcfH and YjjV both have a binuclear metal center. In YcfH, four histidine residues (His-7, His-9, His-130, and His-155) and one aspartate residue (Asp-205) are the ligands of two zinc ions. TatD has only one zinc atom, at a position similar to that of one of the two zinc ions in YcfH/YjjV. Two histidine residues involved in the binding of the metal in YcfH (His-7 and His-9) are not conserved in the TatD sequence. This may be related to the lack of a second bound zinc ion in TatD. The three-dimensional structures of the YcfH orthologs in Thermotoga maritima and Staphylococcus aureus have also been solved (PDB accession numbers 1j6o and 2gzx, respectively). Although the PDB file of the T. maritima ortholog does not include any bound metal, this protein possesses all the conserved amino acid residues indicative of a binuclear metal center. This center is occupied by two nickel ions in the crystalline three-dimensional structure of the S. aureus protein.
The three-dimensional structure of YcfH resembles that of a d-aminoacylase from Alcaligenes faecalis (25). This enzyme, which catalyzes the deacetylation of N-acetyl-d-amino acids, contains two nearby metal-binding sites (called α and β), possibly accounting for an activation/attenuation mechanism (26). AZn2+ ion is always tightly bound to the β site. Additional Zn2+, Cd2+, or Cu2+ at the α site inhibits the activity of the enzyme. Ni2+, Co2+, Mg2+, Mn2+, or Ca2+ were reported to have no effect. Thus, the d-aminoacylase activity is negatively regulated by excess zinc. d-Aminoacylases extracted from Alcaligenes denitrificans or Alcaligenes xylosoxydans behave the same way (27, 28).
Here we have shown by mass spectrometry and atomic absorption spectroscopy that Synechocystis DTD3, a homolog of E. coli YcfH, accommodates various metal ions at two binding sites. DTD3 copurifies with one bound zinc. However, maximum activity is reached if, in addition, a cobalt, a nickel, or a manganese is bound to a second metal site. If the second metal site is occupied by zinc, the enzyme activity is much lower. Therefore, metal ions appear to regulate the activity of DTD3 and possibly those of all the YcfH homologs in a manner close to that already documented with d-aminoacylases. Thermolysin and carboxypeptidase A, representatives of the two large superfamilies of endo- and exoproteases, also show second inhibitory zinc-binding sites within their active site (29, 30).
The phylogenetic tree of proteins sharing extensive sequence homologies with DTD3 is available in the Pfam database. It indicates that E. coli YcfH, YjjV, and TatD proteins are the prototypes of three close protein subsets. TatD-related proteins are present in all eukaryotic genomes sequenced so far. They can also be recognized in many γ-proteobacteria, in Flavobacterium johnsoniae and in two archaea. YjjV homologs are found in many but not all β- and γ-proteobacteria. Finally, the branch containing E. coli YcfH, which includes Synechocystis DTD3, has members in almost all bacteria. Examples that do not display a close homolog to YcfH are only found in γ-proteobacteria belonging to the Alkalilimnicola, Halorhodospira, Carsonella, Xanthomonas, and Xylella genera.
The wide distribution of the DTD1, DTD2, and DTD3 homologs suggests that nearly all cells contain d-aminoacyl-tRNA deacylase activity. This supports the view that the defense of the living world against the invasion of the translational system by d-amino acids is universal, being ensured by at least one out of three distinct types of deacylases. At the present time, only Carsonella ruddii does not possess a gene encoding a protein resembling DTD1, DTD2, or DTD3. C. ruddii is a bacterial endosymbiont lacking numerous genes that are considered essential for life. Therefore, this cell may have achieved an organelle-like status (31). Of note, in the C. ruddii genome, at least 9 aminoacyl-tRNA synthetase genes are missing or altered (32). Possibly as a consequence, the problem raised by an erroneous production of d-aminoacyl-tRNAs is less acute in this bacterium. Finally, the genome of Xylella fastidiosa indicates two DTD3 homologs. However, these homologs are of the YjjV and TatD types. Whether these proteins have deacylase activity is questionable.
In the course of evolution, the early appearance of a d-aminoacyl-tRNA deacylase activity may have been instrumental in fixing the l-amino acid homochirality of the translational machinery. However, under this scenario, the modern deacylases are expected to all derive from a common ancestor. This does not seem to be the case. An alternative scenario would be that several independent deacylases were acquired later. By cooperating with aminoacyl-tRNA synthetases to optimize chiral selective tRNA aminoacylation, these deacylases would have afforded all translational systems a selective advantage. Conversely, the appearance of such deacylase activities may have slowed down the acquisition by aminoacyl-tRNA synthetases of full chiral specificity.
The distribution in the phyla of the three dtd, dtd2, and dtd3-like genes is highly variable. In fact, recent studies indicate that the DTDs may have additional or alternative functions apparently unrelated to the deacylation of a tRNA. Human d-Tyr-tRNATyr deacylase, a homolog of E. coli DTD1, contributes to cell resistance toward d-amino acids (33). However, this protein appears to also be involved in the unwinding of DNA during the initiation of DNA replication (34). In Arabidopsis thaliana, gek1 confers ethanol resistance. The product of this gene, surprisingly, behaves as a DTD2 deacylase (16). Consultation of the Pfam site (in August 2008) revealed that in a few bacteria (Aquifex aeolicus, Magnetococcus sp. MC-1, Kuenenia stuttgartensis, and six δ-proteobacteria), a homolog of DTD3 is genetically fused to a “radical S-adenosylmethionine” domain (35, 36). In fact, as mentioned above, the prevalence of dtd3-like genes is high. In many cells, these genes co-occur with dtd- and/or dtd2-like genes. Such redundancies of the deacylase function may have facilitated an acquisition of alternative or multitasking activities during evolution. Elucidating further the activities of the members in the three DTD1, DTD2, and DTD3 families will certainly help to understand the partition of the deacylase-like proteins in the living world.
Supplementary Material
Acknowledgments
We are greatly indebted to Dr. Jean Houmard for important and helpful advice. We also acknowledge Dr. Gérard Guglielmi and Christian Malosse for their technical assistance in preliminary Synechocystis growth experiments and for mass spectrometry, respectively. We are grateful to Prof. T. Simonson for critical reading of the manuscript.
This work was supported by a grant from the Agence Nationale de la Recherche, 2005–2008.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
Footnotes
The abbreviations used are: IPTG, isopropyl 1-thio-β-d-galactopyranoside; PMSF, phenylmethylsulfonyl fluoride; FA, formic acid; ORF, open reading frame; PDB, Protein Data Bank.
References
- 1.Yang, H., Zheng, G., Peng, X., Qiang, B., and Yuan, J. (2003) FEBS Lett. 552 95–98 [DOI] [PubMed] [Google Scholar]
- 2.Yamane, T., Miller, D. L., and Hopfield, J. J. (1981) Biochemistry 20 7059–7064 [DOI] [PubMed] [Google Scholar]
- 3.Calendar, R., and Berg, P. (1966) Biochemistry 5 1690–1695 [DOI] [PubMed] [Google Scholar]
- 4.Calendar, R., and Berg, P. (1967) J. Mol. Biol. 26 39–54 [DOI] [PubMed] [Google Scholar]
- 5.Soutourina, J., Blanquet, S., and Plateau, P. (2000) J. Biol. Chem. 275 11626–11630 [DOI] [PubMed] [Google Scholar]
- 6.Soutourina, J., Plateau, P., and Blanquet, S. (2000) J. Biol. Chem. 275 32535–32542 [DOI] [PubMed] [Google Scholar]
- 7.Takayama, T., Ogawa, T., Hidaka, M., Shimizu, Y., Ueda, T., and Masaki, H. (2005) Biosci. Biotechnol. Biochem. 69 1040–1041 [DOI] [PubMed] [Google Scholar]
- 8.Sheoran, A., and First, E. A. (2008) J. Biol. Chem. 283 12971–12980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheoran, A., Sharma, G., and First, E. A. (2008) J. Biol. Chem. 283 12960–12970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soutourina, O., Soutourina, J., Blanquet, S., and Plateau, P. (2004) J. Biol. Chem. 279 42560–42565 [DOI] [PubMed] [Google Scholar]
- 11.Soutourina, J., Plateau, P., Delort, F., Peirotes, A., and Blanquet, S. (1999) J. Biol. Chem. 274 19109–19114 [DOI] [PubMed] [Google Scholar]
- 12.Chong, Y. E., Yang, X. L., and Schimmel, P. (2008) J. Biol. Chem. 283 30073–30078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menninger, J. R. (1976) J. Biol. Chem. 251 3392–3398 [PubMed] [Google Scholar]
- 14.Schmitt, E., Mechulam, Y., Fromant, M., Plateau, P., and Blanquet, S. (1997) EMBO J. 16 4760–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferri-Fioni, M. L., Fromant, M., Bouin, A. P., Aubard, C., Lazennec, C., Plateau, P., and Blanquet, S. (2006) J. Biol. Chem. 281 27575–27585 [DOI] [PubMed] [Google Scholar]
- 16.Wydau, S., Ferri-Fioni, M.-L., Blanquet, S., and Plateau, P. (2007) Nucleic Acids Res. 35 930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dardel, F. (1994) Comput. Appl. Biosci. 10 273–275 [DOI] [PubMed] [Google Scholar]
- 18.Vieira, J., and Messing, J. (1991) Gene (Amst.) 100 189–194 [DOI] [PubMed] [Google Scholar]
- 19.van den Bremer, E. T., Jiskoot, W., James, R., Moore, G. R., Kleanthous, C., Heck, A. J., and Maier, C. S. (2002) Protein Sci. 11 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillon, L., Schmitt, E., Blanquet, S., and Mechulam, Y. (2005) Biochemistry 44 15594–15601 [DOI] [PubMed] [Google Scholar]
- 21.Wexler, M., Sargent, F., Jack, R. L., Stanley, N. R., Bogsch, E. G., Robinson, C., Berks, B. C., and Palmer, T. (2000) J. Biol. Chem. 275 16717–16722 [DOI] [PubMed] [Google Scholar]
- 22.Qiu, J., Yoon, J. H., and Shen, B. (2005) J. Biol. Chem. 280 15370–15379 [DOI] [PubMed] [Google Scholar]
- 23.Holm, L., and Sander, C. (1997) Proteins 28 72–82 [PubMed] [Google Scholar]
- 24.Ferri-Fioni, M. L., Schmitt, E., Soutourina, J., Plateau, P., Mechulam, Y., and Blanquet, S. (2001) J. Biol. Chem. 276 47285–47290 [DOI] [PubMed] [Google Scholar]
- 25.Liaw, S. H., Chen, S. J., Ko, T. P., Hsu, C. S., Chen, C. J., Wang, A. H., and Tsai, Y. C. (2003) J. Biol. Chem. 278 4957–4962 [DOI] [PubMed] [Google Scholar]
- 26.Lai, W. L., Chou, L. Y., Ting, C. Y., Kirby, R., Tsai, Y. C., Wang, A. H., and Liaw, S. H. (2004) J. Biol. Chem. 279 13962–13967 [DOI] [PubMed] [Google Scholar]
- 27.Wakayama, M., Yada, H., Kanda, S., Hayashi, S., Yatsuda, Y., Sakai, K., and Moriguchi, M. (2000) Biosci. Biotechnol. Biochem. 64 1–8 [DOI] [PubMed] [Google Scholar]
- 28.Yang, Y. B., Hsiao, K. M., Li, H., Yano, H., Tsugita, A., and Tsai, Y. C. (1992) Biosci. Biotechnol. Biochem. 56 1392–1395 [DOI] [PubMed] [Google Scholar]
- 29.Gomez-Ortiz, M., Gomis-Ruth, F. X., Huber, R., and Aviles, F. X. (1997) FEBS Lett. 400 336–340 [DOI] [PubMed] [Google Scholar]
- 30.Holland, D. R., Hausrath, A. C., Juers, D., and Matthews, B. W. (1995) Protein Sci. 4 1955–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakabachi, A., Yamashita, A., Toh, H., Ishikawa, H., Dunbar, H. E., Moran, N. A., and Hattori, M. (2006) Science 314 267. [DOI] [PubMed] [Google Scholar]
- 32.Tamames, J., Gil, R., Latorre, A., Pereto, J., Silva, F. J., and Moya, A. (2007) BMC Evol. Biol. 7 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, G., Liu, W., Gong, Y., Yang, H., Yin, B., Zhu, J., Xie, Y., Peng, X., Qiang, B., and Yuan, J. (2009) Biochem. J. 417 85–94 [DOI] [PubMed] [Google Scholar]
- 34.Kemp, M., Bae, B., Yu, J. P., Ghosh, M., Leffak, M., and Nair, S. K. (2007) J. Biol. Chem. 282 10441–10448 [DOI] [PubMed] [Google Scholar]
- 35.Frey, P. A., Hegeman, A. D., and Ruzicka, F. J. (2008) Crit. Rev. Biochem. Mol. Biol. 43 63–88 [DOI] [PubMed] [Google Scholar]
- 36.Wang, S. C., and Frey, P. A. (2007) Trends Biochem. Sci. 32 101–110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.