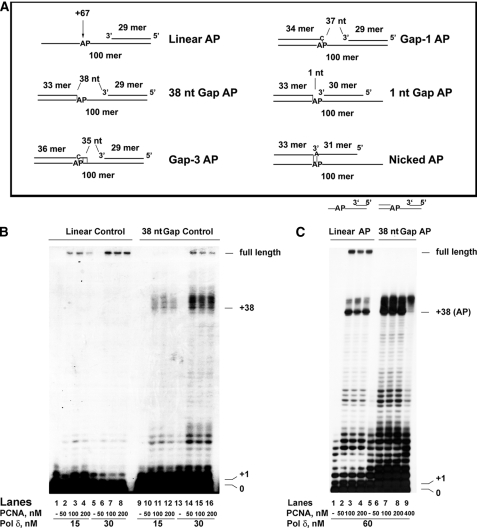
FIGURE 1.
An abasic site immediately upstream of a double-stranded DNA region inhibits the strand displacement activity of DNA polymerase δ. The reactions were performed as described under “Experimental Procedures.” A, schematic representation of the various DNA templates used. The size of the resulting gaps is indicated in nt. The position of the AP site on the 100-mer template strand is indicated relative to the 3′ end. Base pairs in the vicinity of the lesion are indicated by dashes. The size of the gaps (35–38 nt) is consistent with the size of ssDNA covered by a single RP-A molecule, which has to be released during Okazaki fragment synthesis when the DNA pol is approaching the 5′-end of the downstream fragment. When the AP site is covered by the downstream terminator oligonucleotide (Gap-3 and Gap-1 templates) the nucleotide placed on the opposite strand is C to mimic the situation generated by spontaneous loss of a guanine or excision of an oxidized guanine, whereas when the AP site is covered by the primer (nicked AP template), the nucleotide placed on the opposite strand is A to mimic the most frequent incorporation event occurring opposite an AP site. B, human PCNA was titrated in the presence of 15 nm (lanes 2–4 and 10–12) or 30 nm (lanes 6–8 and 14–16) recombinant human four subunit DNA pol δ, on a linear control (lanes 1–8) or a 38-nt gap control (lanes 9–16) template. Lanes 1, 5, 9, and 13, control reactions in the absence of PCNA. C, human PCNA was titrated in the presence of 60 nm DNA pol δ, on a linear AP (lanes 2–4) or 38-nt gap AP (lanes 6–9) template. Lanes 1 and 5, control reactions in the absence of PCNA.