Abstract
Mycobacterium tuberculosis truncated hemoglobin, HbN, is endowed with a potent nitric-oxide dioxygenase activity and has been found to relieve nitrosative stress and enhance in vivo survival of a heterologous host, Salmonella enterica Typhimurium, within the macrophages. These findings implicate involvement of HbN in the defense of M. tuberculosis against nitrosative stress. The protein carries a tunnel system composed of a short and a long tunnel branch that has been proposed to facilitate diatomic ligand migration to the heme and an unusual Pre-A motif at the N terminus, which does not contribute significantly to the structural integrity of the protein, as it protrudes out of the compact globin fold. Strikingly, deletion of Pre-A region from the M. tuberculosis HbN drastically reduces its ability to scavenge nitric oxide (NO), whereas its insertion at the N terminus of Pre-A lacking HbN of Mycobacterium smegmatis improved its nitric-oxide dioxygenase activity. Titration of the oxygenated adduct of HbN and its mutants with NO indicated that the stoichiometric oxidation of protein is severalfold slower when the Pre-A region is deleted in HbN. Molecular dynamics simulations show that the excision of Pre-A motif results in distinct changes in the protein dynamics, which cause the gate of the tunnel long branch to be trapped into a closed conformation, thus impeding migration of diatomic ligands toward the heme active site. The present study, thus, unequivocally demonstrates vital function of Pre-A region in NO scavenging and unravels its unique role by which HbN might attain its efficient NO-detoxification ability.
Unlike many pathogens that are overtly harmful to their host, Mycobacterium tuberculosis can persist for years within humans in a clinically latent state. The success of tubercle bacillus to establish long term persistent infection within the human host lies in its ability to survive and resist hazardous environment of its intracellular niche. Early infection events involve entry and multiplication within the bacteriostatic environment of macrophages (1, 2), where an inducible nitric-oxide synthase generates copious amounts of nitric oxide (NO), which plays an important role in the host defense against microbial pathogens (3, 4) and restricts their growth and survival by inhibiting key enzymes such as the terminal respiratory oxidases (5) and iron-sulfur centers of key enzymes such as aconitase (6, 7). In addition, NO combines with superoxide produced by respiring cells to form the highly oxidizing agent peroxynitrite (8). An efficient NO scavenging system, therefore, is required by microbial pathogens to cope with NO poisoning during their intracellular regime.
M. tuberculosis has evolved resistance mechanisms by which toxic effects of NO and nitrosative stress can be evaded. One of the unique defense mechanisms by which it can protect itself from reactive nitrogen species relies on the oxygenated form of a group I truncated hemoglobin, HbN, which very efficiently converts NO into harmless nitrate (9–11). Studies conducted in our laboratory have shown that HbN carries a potent O2-dependent NO dioxygenase (NOD)3 activity and protects its host from nitrosative stress (11) and microbicidal activities of macrophages (12). It has also been demonstrated that disruption of glbN gene, encoding HbN, in Mycobacterium bovis bacillus Calmette-Guérin causes a dramatic reduction in the NO-consuming activity of stationary phase cells, resulting in marked NO-induced inhibition of aerobic respiration relative to the wild type cells (10). Overall, these studies strongly support the NO-scavenging and detoxification roles of HbN, which may be vital for in vivo survival and pathogenicity of M. tuberculosis.
X-ray crystallographic and computational studies have highlighted the relationship between structure and NOD function of key residues in M. tuberculosis HbN. The protein hosts a two-branched protein matrix tunnel system (13, 14) and has evolved a novel dual-path mechanism to drive migration of O2 and NO to the distal heme cavity where a ligand-induced conformational change regulates the opening of the tunnel branch via phenylalanine (Phe(E15)) residue that acts as a gate for ligand entry (15). Extended molecular dynamics (MD) simulations showed that the essential motions of the protein backbone and the positioning of Phe(E15) gate changes significantly after O2 binding, which facilitates access of a diatomic ligand (NO) to the active site to carry out NO detoxification (16, 17). Finally, a novel pathway has been identified for the egression of nitrate anion (18).
HbN homologs are less represented in living organisms as compared with its group II molecular relative HbO. A variety of physiological functions are displayed by HbN-type truncated hemoglobins (trHbs). For example, Chlamydomonas eugametos HbN has been implicated in photosynthesis (19, 20), whereas the probable function of Paramecium caudatum HbN is O2 supply to mitochondria (21), and HbN of the cyanobacterium Nostoc commune may be part of a microaerobically functioning electron transfer system (22). At present, M. tuberculosis HbN is the only type I truncated hemoglobin for which a potent NOD activity has been ascribed. Recent genome data from various mycobacterial species indicate that HbN-type trHbs exhibit 70–80% sequence similarity (21), which a priori suggests a common globin fold and a similar biological function. Nevertheless, the HbN counterpart in Mycobacterium smegmatis displays severalfold lower NOD activity (23) despite having conserved features for the truncated globin fold and close homology with M. tuberculosis HbN. In fact, residues suggested to play a crucial role for ligand migration through the protein matrix are fully conserved (11). The critical question that emerges now is how M. tuberculosis HbN is able to achieve an efficient NO detoxification ability and what aspects of its structure and/or dynamics contribute to this property.
This study aims to understand the molecular basis of the differences in the NO-metabolizing ability of HbN from M. tuberculosis and M. smegmatis. The results demonstrate a vital contribution of Pre-A helical region in regulation of NOD activity, unraveling a unique mechanism that contributes to modulate the highly efficient NO detoxification activity of HbN.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Culture Conditions—Escherichia coli strains JM109 and BL21DE3 were used routinely for the cloning and expression of recombinant genes. A flavohemoglobin deficient (Δhmp) strain of E. coli (RB9060) has been described earlier (11, 12). Bacterial cultures were grown in Luria-Bertani (LB) or Terrific Broth (containing 24 g of yeast extract, 12 g of Bacto-Tryptone, 12.54 g of K2HPO4, 2.31 g of KH2PO4) medium at 37 °C at 180 rpm unless mentioned otherwise. When required, ampicillin and kanamycin (Sigma) were added at a concentration of 100 and 30 μg/ml, respectively. Plasmids pBluescript (Stratagene) and pET28C (Novagen) were used for cloning and expression of recombinant genes as described earlier (11, 12). Recombinant plasmids pRPN (11) and pSGN (23) were utilized as a source of the HbN-encoding gene of M. tuberculosis and M. smegmatis, respectively. The oligonucleotides were custom-synthesized by Integrated DNA Technologies Inc. Nitric oxide (98.5%) was obtained from Sigma-Aldrich, and saturated NO was prepared as mentioned previously (24).
Cloning, Expression, and Purification of Pre-A Mutants of HbN—Pre-A mutants of HbN were generated using the PCR approach. For the deletion of Pre-A motif and incorporation of mutation within the N-terminal region, forward and reverse primers were designed on the basis of nucleotide sequence of glbN gene. A list of primers used for the construction of various HbN mutants is given in supplemental Table S1. PCR amplified genes encoding HbN mutants were cloned at NdeI-BamHI site of pET28c and expressed under T7 promoter as described previously (11). Recombinant HbN and its mutant proteins were purified from the cell lysate of E. coli using metal affinity chromatography following the standard procedure.
Circular Dichroism (CD) Analysis—Near UV CD spectra of various hemoglobins were monitored at a protein concentration of 33 μm (0.428 mg/ml) in 50 mm Tris-HCl, pH 7.5, at 25 °C. Ten spectra were averaged for each sample at a bandwidth of 2 nm and a scan speed of 100 nm/min. A path length of 1 mm was used for far-UV (250-190 nm) spectral measurement.
Heme Assays—Total cellular heme content was determined as described previously (11). Briefly, ∼2–3 × 109 cells were washed by centrifugation with minimal salt medium (60 mm K2HPO4, 33 mm KH2PO4, 7.6 mm (NH4)2SO4, and 1.7 mm sodium citrate) and were suspended in 0.6 ml of alkaline pyridine reagent containing 2.2 m pyridine and 0.1 n NaOH and lysed with a 30-s sonic burst with a microprobe sonicator. The resulting lysate was then clarified by centrifugation for 10 min at 25,000 × g to remove insoluble debris. Heme concentration was calculated from absorption difference at 556 and 539 nm for the dithionite-reduced and ferricyanide-oxidized sample. The expression level of HbN in the cell was calculated after subtracting the heme content of control cells not expressing any hemoglobin.
Titration of Oxygenated Adducts of HbN and HbN Mutants with NO—Fully oxygenated forms of HbN and HbN mutants were prepared by exposing the deoxygenated protein sample of HbN to air. Absorption spectra of oxygenated HbN were recorded that gave a specific Soret peak at 415 nm and two peaks, α and β, at 540 and 570 nm, very similar to the oxyform of hemoglobin. With a gas tight Hamilton syringe, NO (5 μm) was sequentially added to the oxygenated hemoglobin (40 μm), and absorption spectra were recorded after each addition to convert the oxyhemoglobin into the oxidized form. Profiles of NO-induced oxidation of HbN mutants were compared with the wild type protein.
NO Uptake Assay—NO consumption activity of cells or purified protein was monitored polarographically following the published procedures (25, 26). NO consumption buffer assay contained 60 mm K2HPO4, 33 mm KH2PO4, 7.6 mm (NH4)2SO4, 1.7 mm sodium citrate, 10 mm glucose, and 200 μg/ml chloramphenicol. NO uptake rate of oxygen bound HbN and HbNdelN was measured from the slope of curving traces recorded in the presence of specified concentration of NO (30 μm) following the established protocols (25, 26). HbN catalyzed NO uptake was calculated from three independent measurements and was corrected for background rate of NO decomposition recorded in the control set in the absence of any protein.
Molecular Dynamics Simulations—The dynamical behavior of oxygenated forms of HbN and HbN mutants was examined by means of extended molecular dynamics (MD) simulations, covering a total of 0.5 μs. After our previous studies (16), the x-ray crystallographic structure of wild type M. tuberculosis HbN (Protein Data Bank entry 1idr, chain A at 1.9 Å resolution) was used as starting point for MD simulations (13). Because of the lack of x-ray structures for M. smegmatis HbN, a structural model was built up by a homology model using Swiss-Model (27), taking advantage of the high sequence similarity existing between M. tuberculosis and M. smegmatis HbNs. Simulations were also performed for M. tuberculosis HbN without the Pre-A motif (HbNdelN) and M. smegmatis HbN with the Pre-A motif (HbNextN). Furthermore, an additional simulation was also performed for the reconstituted M. tuberculosis HbN obtained by adding the Pre-A region to the equilibrated HbNdelN structure. In all cases simulations were performed using the same protocol adopted in our previous studies (16, 17). Briefly, the enzyme was immersed in a pre-equilibrated octahedral box of TIP3P (28) water molecules. The final systems contained the protein and around 8600 water molecules (∼ 28,270 atoms). The system was simulated in the NPT ensemble using periodic boundary conditions and Ewald sums for treating long range electrostatic interactions (with the default Amber-9 parameters). The role of Phe(E15) gate residue in the long branch of the tunnel was investigated as described previously (16). All simulations were performed with the parmm99 force field of the Amber-9 package (29) and employing heme parameters developed in previous works (15).
The dynamical behavior of HbN and its mutants was explored by means of essential dynamics (30). To this end, residues 1–15 were excluded as this region is very flexible and would prevent the analysis of the essential motions of the protein core. The backbone atoms in the rest of residues were retained and used to superpose the structures sampled in the central part of the trajectories (20–60 ns). Positional covariance matrixes of the backbone atoms were built up and diagonalized. The eigenvectors define the type of essential motions of the backbone, and the eigenvalues determine how much of the positional variance in the trajectory is explained by each eigenvector.
Finally, the existence of the tunnels in the protein matrix was investigated by means of implicit ligand sampling (ILS) calculations (31), which have been used to identify gas migration pathways inside proteins (32–34). To this end, a set of 10,000 snapshots were taken from every trajectory and enclosed in a grid (0.5 Å spacing) used to place a probe NO molecule. The size of the grid was chosen to fully enclose the protein core and the space corresponding to the layer of surrounding solvent molecules. Parameters for the NO molecule were taken from Cohen et al. (34). 50 orientations of the NO molecule were considered to determine the free energy at each grid point.
RESULTS
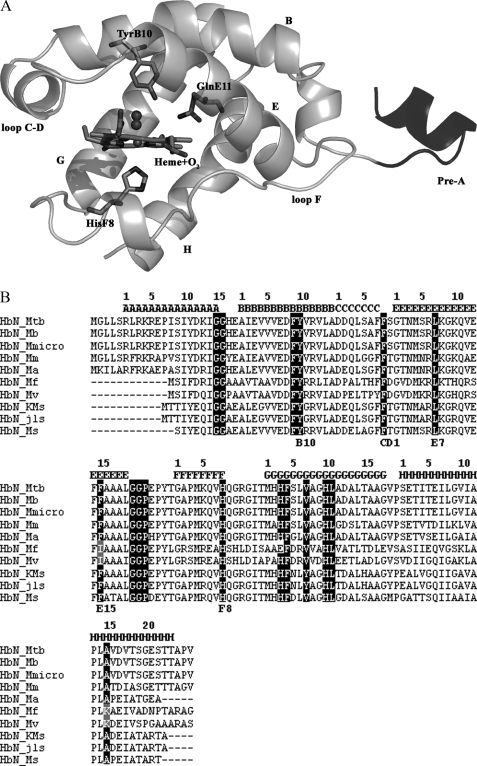
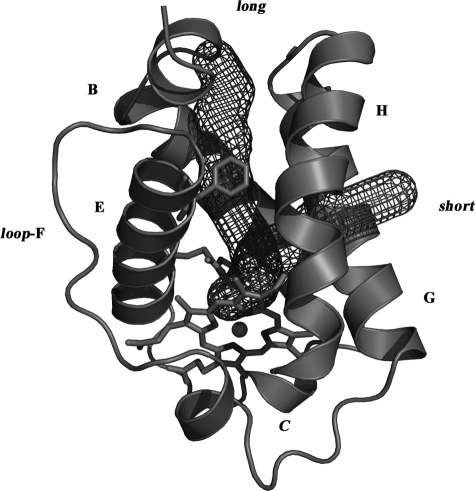
Structural Features of the Pre-A Motif of Mycobacterial HbN— Available mycobacterial genome data suggest that truncated hemoglobins, HbN and HbO, are present in majority of mycobacterial species. Unlike type II-truncated hemoglobin, HbO, which is widely present in various bacteria and plants, occurrence of HbN-type truncated hemoglobin is restricted to only a few species of bacteria and lower eukaryotes; however, the majority of mycobacterial species carry truncated hemoglobin, HbN. Structure-based sequence alignment of different mycobacterial HbNs (Fig. 1B) indicates that the most relevant regions for the stabilization of the truncated globin fold are basically conserved. Nevertheless, HbN found in many fast growing, non-pathogenic species of mycobacterium, e.g. Mycobacterium sp. KMS, Mycobacterium sp. JLS, M. flavescens, M. vanbaalenii, M. smegmatis, etc., lack the 12-residue N-terminal Pre-A motif observed in the x-ray structure of M. tuberculosis HbN (Ref. 13; Fig. 1A) and appears to be present in many slow growing species of mycobacterium such as M. bovis, Mycobacterium avium, Mycobacterium microti, Mycobacterium marinum, etc. that are associated with disease of various organisms. This region, which contains a highly polar sequence motif (Arg-Leu-Arg-Lys-Arg), also distinguishes it from the homologous HbNs present in protozoan, algal, and cyanobacterial species (supplemental Fig. S1). The x-ray structure of M. tuberculosis HbN showed that the Pre-A motif does not contribute significantly to the structural integrity of the protein, as it protrudes out of the compact globin fold. Interestingly, the HbN of most of the slow-growing pathogenic mycobacteria carries this highly charged sequence motif, which is lacking in HbN of many fast growing mycobacteria (Fig. 1B). No structural or functional role has been attributed to this region of HbN at present.
FIGURE 1.
A, structure of M. tuberculosis HbN; structural feature of M. tuberculosis HbN showing the Pre-A motif protruding out of the 2-over-2-globin fold. Key residues and organization of helices and loops are indicated. The amino acid sequence of Pre-A region is shown, and charged residues are marked. B, structure-based sequence alignment of mycobacterial HbN; the globin fold topological positions are marked at the top of alignment following the recent notation given by Vuletich and Lecomte (37). Conserved residues within the truncated hemoglobin family are highlighted, and relevant residues for heme coordination and ligand binding are marked. MTB, M. tuberculosis; Mb, M. bovis; Mmicro, M. microti; Mm, M. marinum; Ma, M. avium; KMS, Mycobacterium sp. KMS; JLS, Mycobacterium sp. JLS; Mf, M. flavescens; Mv, M. vanbaalenii; Ms, Mycobacterium smegmatis. M. tuberculosis, M. bovis, M. microti, M. marinum, and M. avium are slow growing species of mycobacterium. Fast growing species of mycobacteria are Mycobacterium sp. KMS, Mycobacterium sp. JLS, M. flavescens, M. vanbaalenii, and M. smegmatis.
Deletion of the Pre-A Motif from M. tuberculosis HbN Reduces Its Ability to Resist NO Toxicity—To understand the functional relevance of Pre-A motif, this region was deleted from the N terminus of M. tuberculosis HbN, and the modified protein (HbNdelN) was expressed in E. coli. The overexpression of HbNdelN imparted a reddish tinge to the recombinant E. coli, suggesting that the mutant HbN is capable of attaining the truncated globin fold and binding heme. SDS-PAGE analysis of these cells confirmed the presence of a 13-kDa protein corresponding to the expected size of Pre-A-deleted HbN. Spectral analysis of the cell lysate carrying HbNdelN indicated that it is present predominantly in the oxygenated form. O2 and CO binding properties of HbNdelN were similar to the native HbN (data not shown). Interestingly, when HbNdelN was expressed in an NO-sensitive hmp mutant of E. coli and M. smegmatis, it displayed a nearly 7-fold lower NO uptake activity in comparison to isogenic cells carrying native HbN, although the expression level of HbN protein (as determined by the total heme content of cells) and the O2 uptake activity of these cells were comparable (Table 1). These findings suggest that the Pre-A region may be necessary for the optimal NOD function of M. tuberculosis HbN.
TABLE 1.
NO uptake by recombinant E. coli and M. smegmatis carrying Pre-A mutants of HbN Mtb is designated for M. tuberculosis, and Ms is for M. smegmatis. HbNdelN carries a deletion of Pre-A motif in M. tuberculosis HbN, and HbNextN carries the addition of the Pre-A region at the N terminus of M. smegmatis HbN. Data represent the average of five independent sets of experiments for E. coli and three independent sets of M. smegmatis.
| Strains | Heme content | NO consumption | O2 uptake |
|---|---|---|---|
| pmol of heme/1010 cells | nmol of NO heme–1s–1 | μmol/min/1010 cells | |
| E. coli RB9060 (Δhmp) | 2.8 ± 0.4 | 0.04 ± 0.0 | 5.4 ± 0.76 |
| E. coli RB9060 (Mtb HbN) | 36.5 ± 6.1 | 16.8 ± 3.2 | 6.3 ± 0.88 |
| E. coli RB9060 (Mtb HbNdelN) | 42.2 ± 6.6 | 2.8 ± 1.3 | 6.5 ± 0.69 |
| E. coli RB9060 (Ms HbN) | 31.7 ± 5.4 | 3.8 ± 1.6 | 6.1 ± 0.73 |
| E. coli RB9060 (Ms HbNextN) | 38.1 ± 4.6 | 8.9 ± 2.2 | 6.0 ± 0.77 |
| M. smegmatis (wild type) | 0.85 ± 0.11 | 0.0 ± 0.0 | 2.8 ± 0.14 |
| M. smegmatis (Mtb HbN) | 14.80 ± 2.61 | 27.82 ± 3.5 | 3.7 ± 0.32 |
| M. smegmatis (Mtb HbNdelN) | 17.44 ± 1.82 | 3.43 ± 1.1 | 3.9 ± 0.45 |
| M. smegmatis (Ms HbN) | 16.60 ± 2.1 | 4.9 ± 1.5 | 4.1 ± 0.36 |
| M. smegmatis (Ms HbNextN) | 15.11 ± 2.9 | 11.8 ± 1.3 | 4.3 ± 0.55 |
Integration of Pre-A Motif at the N Terminus of Pre-A Lacking HbN of M. smegmatis—To further study the implications of Pre-A on NO uptake function of protein, we engineered this motif of M. tuberculosis HbN at the N terminus of M. smegmatis HbN that lacks a Pre-A region. Expression of Pre-A-carrying M. smegmatis HbN (HbNextN) in the hmp mutant of E. coli displayed 2-fold higher NOD activity over the native M. smegmatis HbN, although the NOD activity of HbNextN was still 1.5-fold lower than the M. tuberculosis HbN (Table 1). Wild type M. smegmatis did not show any measurable NOD activity under aerobic condition; however, cloning and expression of M. smegmatis HbN in these cells resulted in the appearance of a low but distinct level of NOD activity very similar to the isogenic cells expressing HbNdelN mutant (Table 1). The NO uptake activity of cells increased 2–2.5-fold when the Pre-A-carrying mutant of M. smegmatis HbN (HbNextN) was expressed in M. smegmatis using the same expression plasmid. The level of heme content and oxygen uptake rate of these cells carrying a different HbN expression plasmid were nearly the same (Table 1). Overall, the results demonstrate that the Pre-A motif plays a critical role in modulating the NOD function of HbN, although additional factors (possibly reflecting the effect of specific point mutations within the two species of HbN) contribute to the modulation of the functional efficiency of mycobacterial HbNs.
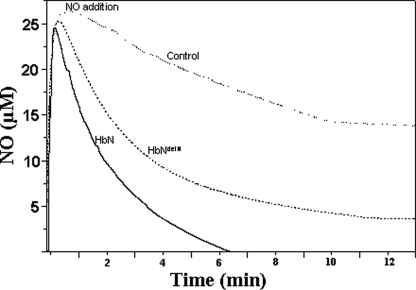
NO Scavenging by Pre-A-deleted HbN of M. tuberculosis HbN in Vitro—We further checked how deletion of Pre-A from HbN would affect its NO scavenging activity in vitro to rule out the involvement of any cellular factors in regulating the effect of Pre-A region on the NOD activity of HbN. Because catalytic NO scavenging would require reduction of the ferric heme iron, a ferredoxin reductase was used as an artificial reductase system to determine NO uptake by the oxygenated form of wild type HbN and HbNdelN in vitro. With no HbN present, the addition of 30 μm NO to the control set resulted in slow autooxidation of NO, and the NO signal diminished completely and reached the base line in 30 min. The presence of 15 μm wild type oxy-HbN resulted in rapid removal of NO from the reaction mixture, and NO signal reached base line within 4–5 min after NO addition. However, when the oxygenated form of Pre-A-deleted HbN was used in place of wild type HbN under similar conditions, the removal of NO was significantly slower and reached base line after 12–14 min (Fig. 2). The rate of NO uptake by HbN (216 ± 10.6 nmol/min/mg, calculated from three sets of experiments) was found nearly 6-fold higher in comparison to HbNdelN (35.4 ± 4.1 nmol/min/mg) under similar conditions. These results further substantiated the vital contribution of Pre-A in regulating the NO scavenging ability of M. tuberculosis HbN.
FIGURE 2.
NO consumption activity of HbN and Pre-A-deleted HbN in vitro. Consumption of NO by oxygenated form of HbN and HbNdelN was measured in vitro by NO electrode (World Precision Instrument) by recording the rate of NO removal after the addition of NO (30 μm) to 2 ml of 50 mm potassium phosphate buffer, pH 7.2, containing 100 μm NADH, 1 μm ferredoxin reductase (Sigma), and 15 μm wild type and mutant HbN. Buffer without any HbN protein was taken as the control.
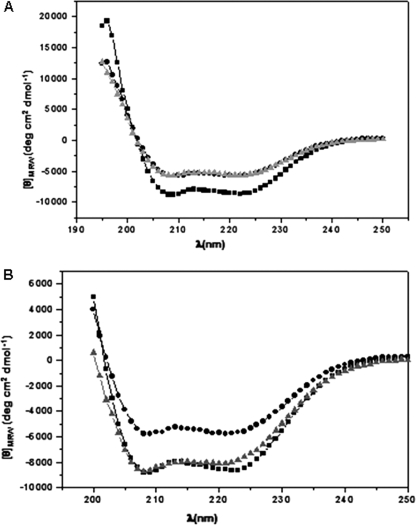
Circular Dichroism Spectra of Pre-A Mutants of HbN—To check whether the deletion or addition of Pre-A has any effect on overall protein structure, the CD spectra was recorded. Far-UV spectra of M. tuberculosis and M. smegmatis HbN and their Pre-A mutants displayed two strong negative bands at θ209 nm and θ222 nm, which indicates similar structures having high amount of α-helical character. However, the content of random coil decreased when Pre-A was deleted from M. tuberculosis HbN (HbNdelN) and increased when Pre-A region was added to M. smegmatis HbN (HbNextN) (Fig. 3), but no major change in overall structure of protein was observed, suggesting that Pre-A region does not contribute to the structural integrity of protein.
FIGURE 3.
Structural properties of Pre-A-deleted and Pre-A-carrying mutants of HbN. A, the near-UV CD spectra of HbN of M. tuberculosis (▪), M. smegmatis (•), and M. tuberculosis HbN mutant deleted in Pre-A region (▴). B, the near-UV CD spectra of HbN of M. tuberculosis (▪), M. smegmatis (•), and M. smegmatis HbN-carrying Pre-A region of M. tuberculosis HbN (▴).
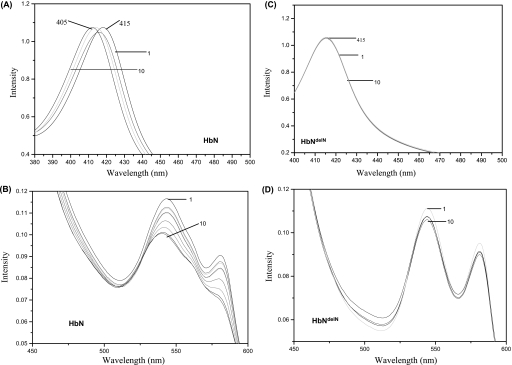
NO Oxidation by Pre-A Lacking HbN in the Presence of Oxygen—Pre-A mutants of HbN displayed more or less similar O2 binding properties and were able to generate a stable oxy form displaying a similar Soret peak at 415 nm and α and β peaks at 545 and 570 nm. Because NO scavenging ability of hemoglobins may also depend on their efficiency to oxidize NO after O2 binding, we compared a pattern of NO oxidation by O2-bound HbN and its Pre-A-lacking mutant, HbNdelN. The addition of NO (5 μm) to oxygenated HbN (40 μm) resulted in the appearance of a partially oxidized spectrum (Fig. 4, A and B). Repeated addition of NO solution to this sample resulted in a fully oxidized spectrum of HbN that was evident from the shifting of Soret peak of oxyHbN at 415–405 nm. When Pre-A-deleted HbN was exposed to NO under similar conditions, oxidized spectrum did not appear immediately after similar additions of NO and changed very slowly after 30–40 min of exposure (Fig. 4, C and D), which indicates slow oxidation of NO by HbN after Pre-A removal.
FIGURE 4.
Oxidation of O2-bound HbN and Pre-A-deleted HbN by NO. The ligand-free deoxygenated form of HbN and HbN mutant was exposed to air to oxygenate the protein sample. Spectra of the oxygenated HbN (A and B) and HbNdelN (C and D) were recorded before and after sequential addition of saturated solution of NO (5 μm) to the protein (40 μm). The first (1) and last additions (10) of NO have been designated. Fully oxidized spectra of wild type HbN has been obtained after 9 addition of 5 μm NO.
MD Simulations of Pre-A Mutants of HbN—The effect of the Pre-A region on the dynamical behavior of native and mutated forms of M. tuberculosis and M. smegmatis HbNs was examined by means of MD simulations.
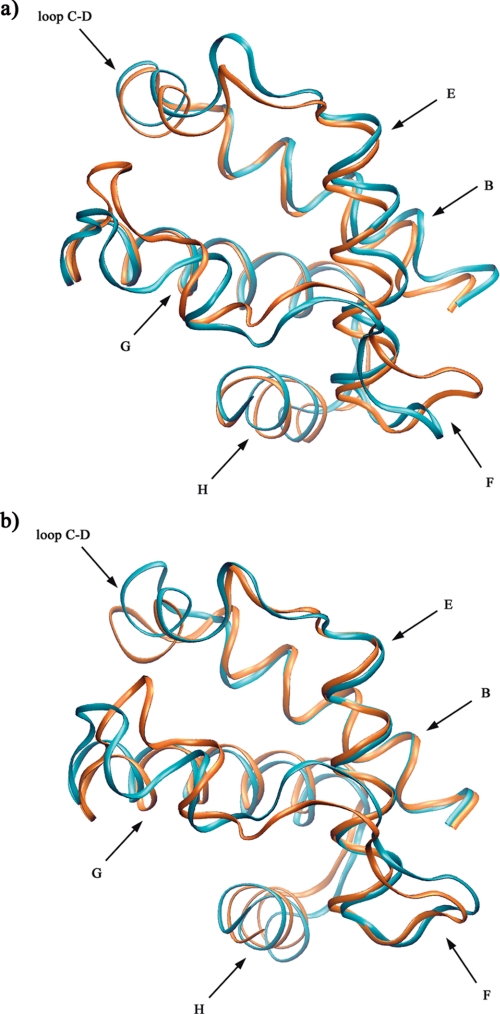
Because of the high sequence similarity found between M. tuberculosis and M. smegmatis HbNs (close to 80%), homology modeling was used to build up a structural model of M. smegmatis HbN. The two branches of the tunnel observed in the x-ray crystallographic structure of M. tuberculosis HbN are well preserved in the model structure of M. smegmatis HbN (see Fig. 5), thus reflecting the high degree of conservation for the residues that delineate the walls of the tunnel. Specifically, it is worth noting that the residues expected to have a key functional role for ligand migration in M. tuberculosis HbN, such as the gate Phe(E15), which controls the opening of the tunnel long branch, or the molecular switch formed by the pair Tyr(B10)-Gln(E11), which facilitates the opening of the gate upon O2 binding to the heme (see Refs. 15–17 for more details), are retained and properly positioned in the model structure. A similar finding is found for Thr(E10), which has been proposed to guide the egression of the nitrate anion (18). Accordingly, it is reasonable to assume that the molecular basis of the NO detoxification mechanism will be highly similar in HbNs from M. tuberculosis and M. smegmatis.
FIGURE 5.
Organization of tunnel in M. smegmatis HbN. Representation of the long and short branches of the tunnel in the structural model built up for M. smegmatis HbN by homology modeling. Contours were determined using the program CAVER (38).
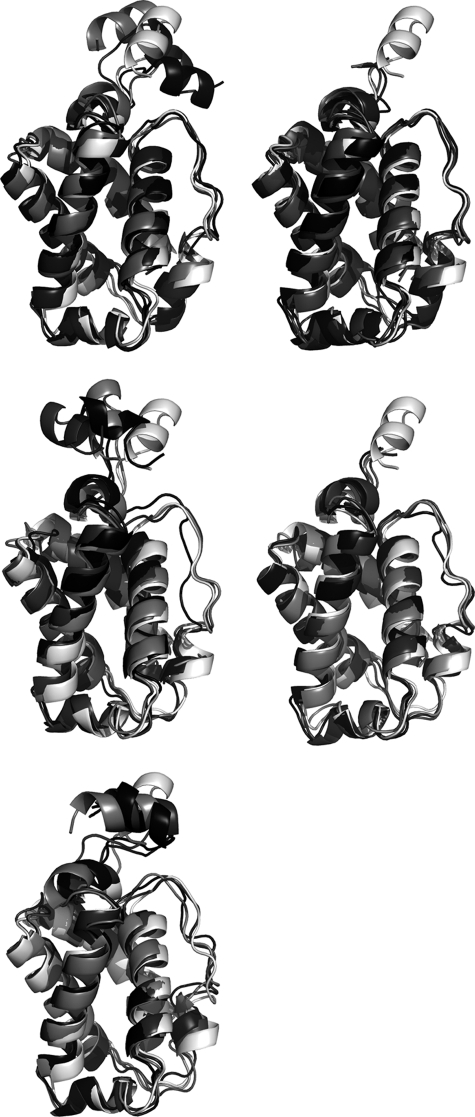
In all cases the stability of the trajectories was confirmed upon inspection of the time profiles of the potential energy and root-mean square deviation determined for the backbone atoms (r.m.s.d.; see supplemental Fig. S2). The stability of the trajectories is supported by the comparison of the superposed protein backbone for the x-ray crystallographic structure (Protein Data Bank entry 1idr) and the snapshots collected at 10, 50, and 100 ns, as noted in Fig. 6, which shows that the largest fluctuations correspond to the Pre-A region and the loop F. Thus, upon exclusion of these regions, the r.m.s.d. between the last snapshot of the trajectories and the x-ray crystallographic structure ranges from 1.3 to 2.3 Å (see Table 2). Moreover, the r.m.s.d. determined for the backbone atoms of the protein core between the average structures of the wild type and mutated forms of HbN varies from 0.6 to 1.5 (see supplemental Table S2). Overall, these analyses indicate that the protein core remains stable along all the trajectories and that the main structural fluctuations in the wild type M. tuberculosis HbN and the mutated M. smegmatis HbNextN are largely due to the Pre-A region, which can be ascribed to the protruding nature of the Pre-A region from the core of the protein.
FIGURE 6.
Superposition of the backbone for representative snapshots collected along the trajectories. Snapshots taken at 10 (light gray), 50 (dark gray), and 100 (black) ns along the trajectories run for the wild type and mutated forms of HbN are superposed with the x-ray crystallographic structure (Protein Data Bank entry 1idr, white). The superposition is shown for the wild type M. tuberculosis HbN (top left) and its Pre-A-deleted mutant (top right), the M. smegmatis trHbN with inserted Pre-A region (middle left) and the wild type M. smegmatis HbN (middle right), and finally, the rebuilt form of M. tuberculosis HbN (bottom).
TABLE 2.
Root-mean square deviations (Å) between the x-ray crystallographic structure of M. tuberculosis trHbN (PDB entry 1idr) and the last snapshot collected along the trajectory, determined by considering the atoms of the whole protein backbone and upon exclusion of the Pre-A region and loop F Mtb is designated for M. tuberculosis, and Ms is for M. smegmatis. HbNdelN carries the deletion of the Pre-A motif in M. tuberculosis HbN, and HbNextN carries the addition of the Pre-A region at the N terminus of M. smegmatis HbN.
| Protein | Whole backbone | Excluding Pre-A and loop F |
|---|---|---|
| Mtb HbN | 3.0 | 1.3 |
| Mtb HbNdelN | 1.9 | 1.8 |
| Mtb HbN (rebuilt) | 3.4 | 2.1 |
| Ms HbN | 2.1 | 1.9 |
| Ms HbNextN | 3.5 | 2.3 |
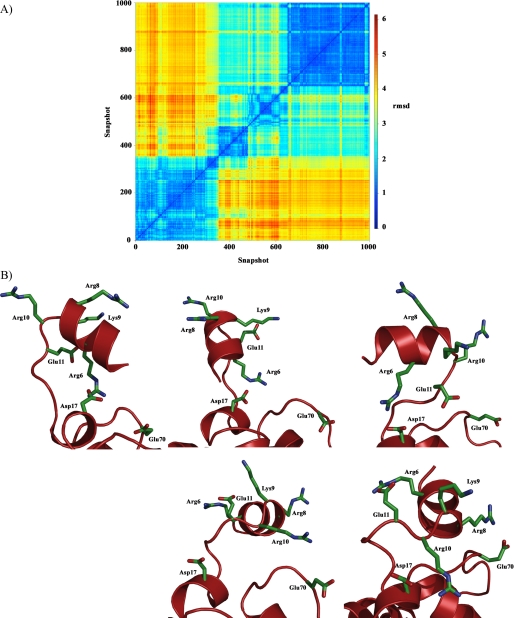
For the wild type M. tuberculosis HbN, the Pre-A segment was found to adopt four main representative orientations along the 100-ns trajectory (see supplemental Figs. S3 and S4 and Table S3), thus reflecting the large flexibility of this region. Even though the salt bridge seen in the x-ray crystallographic structure between Arg-6 and Asp-17 is retained in the two first clusters (sampled in the first 47 ns), this interaction is lost in the rest of the trajectory (Fig. 7). In fact, Arg-6 interacts with Glu-11 in the last part of the trajectory. Overall, the results suggest that transient salt bridges are formed from the balance between electrostatic contacts between charged residues and hydration with water molecules.
FIGURE 7.
Structural flexibility of the Pre-A region in M. tuberculosis HbN. A, plot of cross-r.m.s.d. (Å) determined for backbone atoms between snapshots (taken every 100 ps) collected along the 100-ns trajectory for the M. tuberculosis HbN (cross-r.m.s.d. values were computed using the ptraj module of AMBER and plotted using R software). B, arrangement of charged residues in Pre-A region found in (top left) the x-ray crystallographic structure 1idr and four representative snapshots taken as the closest members to the centroid of the structural clusters depicted above (ordered from left to right and top to bottom; see also Fig. supplemental S4).
The analysis of MD trajectories revealed that the relative movement of protein backbone changes significantly when Pre-A is removed from the protein core (Fig. 8, a and b). A distinct difference in the dynamics of protein structure was observed around helices B and E and the EF loop region. Thus, helices B and E, which largely contribute to delineate the long branch of the tunnel, exhibit larger fluctuations in M. tuberculosis HbN compared with the HbNdelN structure. In fact, the first essential mode in M. tuberculosis HbNdelN involves motions of helices G and H and loop CD (Fig. 8). Overall, Pre-A-deleted HbN displayed a clear cut restriction in the movement of B and E helices as well as the EF loop region that may affect functioning of long tunnel branch and Phe(E15) gate.
FIGURE 8.
Representative snapshots showing protein backbone movement in HbN and Pre-A-deleted HbN. Representation of the essential dynamics results for the wild type HbN (a) and its Pre-A-deleted mutant (b). The distinct motion leading to dynamical fluctuation in the protein backbone and helices (shown in blue and brown colors) is represented using graphical facilities in VMD program (39).
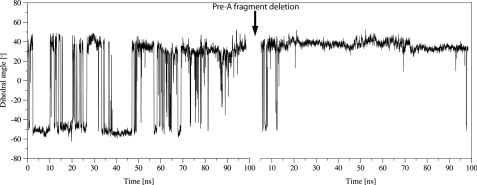
In agreement with these results and our previous findings (15–17), the evolution of the dihedral angle HA-CA-CB-CG of Phe(E15) along the trajectory sampled for M. tuberculosis HbN showed the occurrence of several opening/closing events (Fig. 9; an alternative representation made by using the dihedral angle N-CA-CB-CG is given in supplemental Fig. S5) that modulate the entry of NO toward the active site. Nevertheless, deletion of Pre-A from HbN gives rise to a dramatic change in the protein backbone motion, and the Phe(E15) gate is trapped into a closed conformation along the whole trajectory (Fig. 9), thus blocking easy access of NO toward the active site. Importantly, reconstitution of the wild type M. tuberculosis HbN upon the addition of the Pre-A region to the last sampled structure of the HbNdelN trajectory restored the opening/closing events of Phe(E15) gate (Fig. 10c). At this point it is worth noting that ILS calculations clearly identify the long branch of the tunnel as the lowest energy pathway for ligand (NO) migration along the protein core for either the wild type M. tuberculosis HbN and its Pre-A-deleted mutant (see Fig. 11). Accordingly, although the tunnel long branch is identified as the most feasible migration pathway, the reduction in the NO consumption of the Pre-A-deleted mutant can be attributed to the trapping of the Phe(E15) gate into the closed conformation.
FIGURE 9.
Conformational transition between open and closed states of the Phe(E15) gate of M. tuberculosis HbN. Open and closed states are characterized by the torsional dihedral HA-CA-CB-CG of Phe(E15). After 100 ns of MD of the wild type M. tuberculosis HbN (left), the Pre-A motif was deleted, and the mutant was re-equilibrated and subjected to 100 ns MD (right).
FIGURE 10.
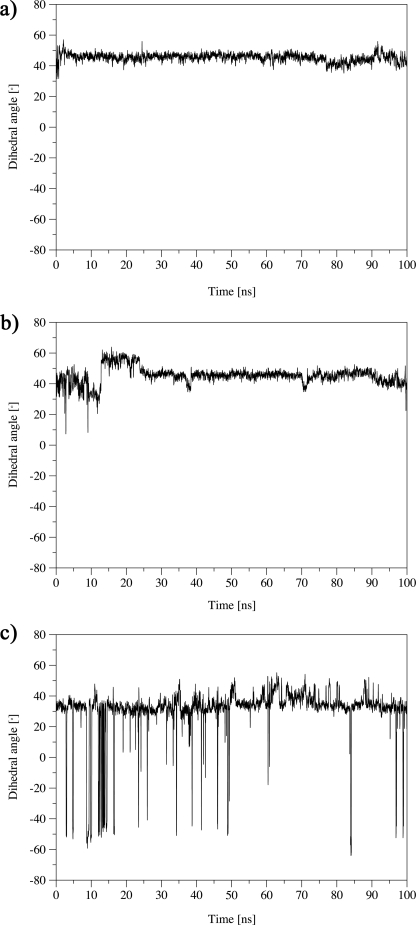
Conformational transition of the Phe(E15) gate in Pre-A mutant of HbN after deletion and reinsertion. Conformational transition between the open and closed states of the Phe(E15) gate of HbN (characterized by the torsional dihedral HA-CA-CB-CG of Phe(E15)) along 100 ns of MD simulations of wild type M. smegmatis HbN (a), its Pre-A-inserted mutant (b), and the rebuilt HbN of M. tuberculosis protein after reinsertion of Pre-A in HbNdelN (c).
FIGURE 11.
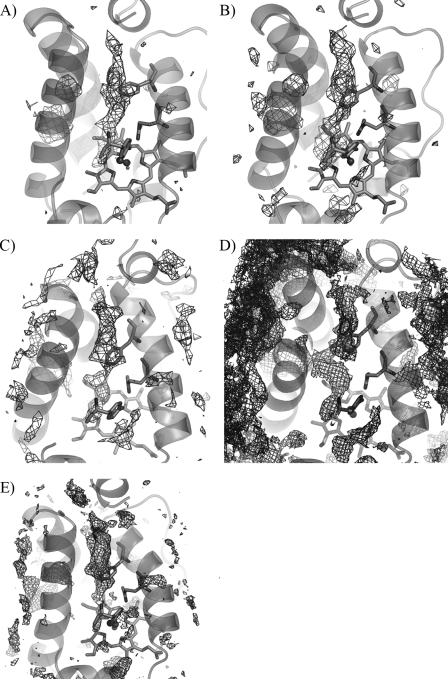
Representation of the lowest energy pathway in wild type and mutated proteins. The most feasible pathways leading from the heme cavity to the protein surface were determined for migration of NO by means of implicit ligand sampling calculations for wild type M. tuberculosis HbN (A), its Pre-A-deleted mutant (B), Pre-A-inserted M. smegmatis HbN (C), wild type M. smegmatis HbN (D), and rebuilt wild type M. tuberculosis HbN (E) (isocontour values of -0.5, 0.8, 1.9 and 4 kT are used for A, B and C, D, and E, respectively). The long branch of the tunnel is found to be the lowest energy pathway in all cases. Only in wild type M. smegmatis HbN are both long and short tunnels detected simultaneously. If the wild type M. tuberculosis HbN is contoured using the level defined for D, the short tunnel is delineated, although there still exists a break in the path. Likewise, if D were contoured at the level of A, no continuous path would have been found.
Mimicking the behavior found for HbNdelN, the 100-ns trajectories sampled for the wild type M. smegmatis HbN and its Pre-A-inserted mutant displayed no relevant transition between open and closed conformations of Phe(E15) (Fig. 10, a and b). Again, ILS computations reveal that the long branch of the tunnel is the lowest energy pathway identified in the Pre-A-extended form of M. smegmatis HbN (see Fig. 11), as was noted before for the wild type M. tuberculosis protein and its Pre-A-deleted mutant. However, it is worth noting that for the wild type M. smegmatis HbN the energy isocontour chosen to delineate the channels leading from the heme cavity to the protein surface identifies both short and long branches of the tunnel (Fig. 11). Accordingly, it can be suggested that the addition of the Pre-A region tends to favor the migration of ligands through the tunnel long branch.
Although differences in specific residues (i.e. point mutations) within the core region of M. tuberculosis and M. smegmatis HbNs can contribute to the reduced conformational transition of Phe(E15) gate, the lack of opening events in the trajectory for M. smegmatis Pre-A-extended HbN might also be affected by the limited flexibility observed for the Pre-A region. Thus, in contrast with the behavior found for M. tuberculosis trHbN, the Pre-A segment in the M. smegmatis mutant adopts two main structural arrangements, the last one being found in the last 80 ns of the trajectory. Moreover, whereas the Arg-6 —Asp-17 salt bridge was lost in the last 50 ns for the M. tuberculosis trHbN, Arg-6 is permanently bound to Glu-17 in the M. smegmatis mutant (see supplemental Figs. S6 and S7). Thus, it can be speculated that the larger flexibility of the side chain of Glu-17 facilitates the interaction with Arg-6, which in turn could limit the global flexibility of the Pre-A region.
DISCUSSION
The involvement of M. tuberculosis HbN in protection from the reactive nitrogen intermediates, particularly NO, has substantially been ascribed to its efficient NOD activity (9–11) that is able to convert toxic NO into innocuous nitrate nearly 15-fold faster than the horse heart myoglobin (10). The molecular mechanism by which the single domain HbN of M. tuberculosis attains this function and the aspects of its structure and dynamics responsible for its enhanced reactivity are not completely known at present and poses an intriguing question yet to be answered. The present study demonstrates a crucial role of the Pre-A motif in modulating the NO-scavenging function of HbN through a novel mechanism.
A Pre-A helical region is absent in HbN of many mycobacterial species as well as HbN from other organisms including the extended N terminus of Chlamydomonas and Pseudomonas (see supplemental Fig. S1) and may not be necessary for the structural integrity of the protein. Deletion of Pre-A motif from M. tuberculosis HbN expressed in both E. coli and M. smegmatis did not change the global protein fold nor significantly affect the O2 binding properties but drastically reduces its NOD activity. Therefore, the Pre-A region seems to be a requirement for efficient NO detoxification of HbN. This is supported by the fact that M. smegmatis HbN, which lacks the Pre-A region, exhibits very low NOD activity and is unable to protect recombinant E. coli from the toxicity of NO and acidified nitrite (23) despite the high degree of sequence homology with M. tuberculosis HbN.
The NO-metabolizing activity of HbN largely depends on efficiency of ligand migration to the heme cavity (9–11), which follows a dual path mechanism for the entry of O2 and NO through the branched tunnel (13, 16, 17). After migration through the tunnel short branch, binding of O2 to the heme cavity facilitates migration of the second ligand, i.e. NO, through the tunnel long branch by triggering the opening of the Phe(E15) gate. Because deletion of Pre-A region impairs the ability of HbN to oxidize NO stoichiometrically both in vivo and in vitro, it seems reasonable to hypothesize that the slow NO uptake and its impaired oxidation with heme-bound O2 is because of limited migration of NO to the heme cavity. Extended MD simulations and ILS results substantiated this hypothesis. A distinct change in protein backbone motion appeared when Pre-A region was deleted from the HbN. The dynamical alteration observed upon O2 binding in M. tuberculosis HbN (16, 17), which mainly involves displacement of B and E helices along with local rearrangements (primarily the side chain of Gln(E11)) near the Phe(E15) gate, is not found in the trajectories run for M. tuberculosis Pre-A-deleted HbN (HbNdelN). ILS results indicate that in the wild type M. tuberculosis HbN and its Pre-A-deleted mutant, the tunnel long branch is identified as the lowest energy migration pathway, thus supporting the suggestion that the reduction in the NO consumption of the mutant can be attributed to the fact that Phe(E15) gate is trapped in the closed conformation. Importantly, the ability of Phe(E15) gate to switch between open and closed states was recovered when the Pre-A region was reinserted into Pre-A-deleted HbN of M. tuberculosis.
In the crystal structure (13) the Pre-A region is folded, making close contacts with G helix and EF loop of HbN. Nevertheless, as found in MD simulations, it is very flexible and adopts different arrangements along the trajectory. Accordingly, it can be speculated that thermal fluctuations of the Pre-A region influence the backbone motion of the protein and in turn affect the conformational state of the Phe(E15) gate and migration of ligands through the tunnel long branch. This unexpected role is also supported by the results found for wild type M. smegmatis HbN, whose NOD activity is several folds lower than its counterpart in M. tuberculosis (23). Interestingly, both experimental and simulation results indicate that simple insertion of Pre-A region does not suffice to fully recover the ligand migration properties of M. smegmatis HbN, suggesting that, apart from Pre-A segment, other differences within the core region of M. tuberculosis and M. smegmatis HbNs also contribute to the NOD function. In particular, it can be hypothesized that the enhanced side chain of Glu-4 in M. smegmatis, which replaces Asp-17 in wild type M. tuberculosis trHbN, could facilitate the maintenance of the salt bridge with Arg-6, which in turn could reduce the global intrinsic flexibility of Pre-A segment and eventually decrease its effect on the dynamics of the protein core. Alternatively, it can also be speculated that mutations could affect the global dynamical “breathing” of the protein matrix, specifically the relative motion of helices B and E, which mainly contribute to delineate the long branch of the tunnel. This interpretation presents conceptual differences with the effect of points mutations on ligand migration in myoglobin (35) where ligand release depends on transient trapping of the ligand into internal cavities.
Recent studies on transcriptional regulation of M. tuberculosis hemoglobins have shown that a significant level of HbN is produced during macrophage infection (36), and its NOD activity is sustained under low oxygen (12). Even though HbN-type truncated hemoglobins possess a tunnel system within the protein matrix similar to that of M. tuberculosis HbN, none of these have been demonstrated to have potent NOD activity. Then, the Pre-A region appears to be a unique structural feature of HbN of some pathogenic mycobacteria. It is unknown whether dynamic movement and folding properties of Pre-A changes under different physiological conditions of cells and regulates NOD activity of HbN to achieve maximum benefit under stress conditions.
Taken together the present study unravels three major findings. First, the Pre-A motif is important for the optimal NO scavenging activity of M. tuberculosis HbN measured upon expression in E. coli and M. smegmatis. Second, swapping of the Pre-A motif onto a Pre-A lacking HbN of M. smegmatis improves its NOD activity but not to the level of M. tuberculosis HbN, which suggests that other factors also contribute to the effect of Pre-A on HbN function. Finally, extended MD simulations point out that the deletion of Pre-A region might alter the protein dynamics and conformational state of the long tunnel gate that restricts passage of NO to the distal heme cavity, thus impairing the ability of HbN for NO detoxification. Overall, these findings unveil an unexpected role of pre-A segment by which M. tuberculosis HbN may carry out NO detoxification efficiently to cope with enhanced level of toxic nitrogen species generated within the intracellular environment.
Supplementary Material
Acknowledgments
Help provided by Sandip Singh during the experimental work is thankfully acknowledged.
This work was supported by the Department of Biotechnology, Government of India, and Council of Scientific and Industrial Research, Spanish Ministerio de Ciencia e Innovación Grants SAF2008-05595-C02-01 and PCI2006-A7-0688, Agencia Nacional de Promoción Científica y Tecnológica Grant PICT 25667, Consejo Nacional de Investigaciones Científicas y Técnicas, Universidad de Buenos Aires, the John Simon Guggenheim Foundation, and the European Union FP7 program (project NOstress).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1–S3.
Footnotes
The abbreviations used are: NOD, nitric-oxide dioxygenase; trHb, truncated hemoglobin; MD, molecular dynamics; ILS, implicit ligand sampling; r.m.s.d., root-mean square deviation.
References
- 1.Fenton, M. J., and Vermeulen, M. W. (1996) Infect. Immun. 64 683-690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fenton, M. J. (1998) Curr. Opin. Hematol. 5 72-78 [DOI] [PubMed] [Google Scholar]
- 3.Chan, J., Xing, Y., Magliozzo, R. S., and Bloom, B. R. (1992) J. Exp. Med. 175 1111-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacMicking, J. D., North, R. J., LaCourse, R., Mudgett, J. S., Shah, S. K., and Nathan, C. F. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5243-5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevanin, T. M., Poole, R. K., Demoncheaux, E. A., and Read, R. C. (2002) Infect. Immun. 70 4399-4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner, P. R., Constantino, G., Szabo, C., and Salzman, A. L. (1997) J. Biol. Chem. 272 25071-25076 [DOI] [PubMed] [Google Scholar]
- 7.Gardner, P. R., Constantino, G., and Salzman, A. L. (1998) J. Biol. Chem. 273 26528-26533 [DOI] [PubMed] [Google Scholar]
- 8.Beckman, J. S., Beckman, T. W., Chen, J., Marshall, P. A., and Freeman, B. A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 1620-1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couture, M., Yeh, S. R., Wittenberg, B. A., Wittenberg, J. B., Ouellet, Y., Rousseau, L., and Guertin, M. A. (1999) Proc. Natl. Acad. Sci. U. S. A., 96 11223-11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouellet, H., Ouellet, Y., Richard, C., Labarre, M., Wittenberg, B. A., Wittenberg, J. B., and Guertin, M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5902-5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pathania, R., Navani, N. K., Gardner, A. M., Gardner, P. R., and Dikshit, K. L. (2002) Mol. Microbiol. 45 1303-1314 [DOI] [PubMed] [Google Scholar]
- 12.Pawaria, S., Rajamohan, G., Gambhir, V., Lama, A., Varshney, G. C., and Dikshit, K. L. (2007) Microb. Pathog. 42 119-128 [DOI] [PubMed] [Google Scholar]
- 13.Milani, M., Pesce, A., Ouellet, Y., Ascenzi, P., Guertin, M., and Bolognesi, M. (2001) EMBO J. 20, 3902-3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milani, M., Pesce, A., Ouellet, Y., Dewilde, S., Friedman, J., Ascenzi, P., Guertin, M., and Bolognesi, M. (2004) J. Biol. Chem. 279 21520-21525 [DOI] [PubMed] [Google Scholar]
- 15.Crespo, A., Martí, M. A., Kalko, S. G., Morreale, A., Orozco, M., Gelpí, J. L., Luque, F. J., and Estrin, D. A. (2005) J. Am. Chem. Soc. 127 4433-4444 [DOI] [PubMed] [Google Scholar]
- 16.Bidon-Chanal, A., Martí, M. A., Crespo, A., Milani, M., Orozco, M., Bolognesi, M., Luque, F. J., and Estrin, D. A. (2006) Proteins 64 457-464 [DOI] [PubMed] [Google Scholar]
- 17.Bidon-Chanal, A., Martí, M. A., Estrin, D. A., and Luque, F. J. (2007) J. Am. Chem. Soc. 129 6782-6788 [DOI] [PubMed] [Google Scholar]
- 18.Martí, M. A., Bidon-Chanal, A., Crespo, A., Yeh, S. R., Guallar, V., Luque, F. J., and Estrin, D. A. (2008) J. Am. Chem. Soc. 130 1688-1693 [DOI] [PubMed] [Google Scholar]
- 19.Couture. M., Chamberland, H., St-Pierre, B., Lafontaine, J., and Guertin, M. (1994) Mol. Gen. Genet. 243 185-197 [DOI] [PubMed] [Google Scholar]
- 20.Das, T. K., Couture, M., Lee, H. C., Peisach, J., Rousseau, D. L., Wittenberg, B. A., Wittenberg, J. B., and Guertin, M. (1999) Biochemistry 38 15360-15368 [DOI] [PubMed] [Google Scholar]
- 21.Wittenberg, J. B., Bolognesi, M., Wittenberg, B. A., and Guertin, M. (2002) J. Biol. Chem. 277 871-874 [DOI] [PubMed] [Google Scholar]
- 22.Hill, D. R., Belbin, T. J., Thorsteinsson, M. V., Bassam, D., Brass, S., Ernst, A., Boger, P., Paerl, H., Mulligan, M. E., and Potts, M. (1996) J. Bacteriol. 178 6587-6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lama, A., Pawaria, S., and Dikshit, K. L. (2006) FEBS Lett. 580 4031-4041 [DOI] [PubMed] [Google Scholar]
- 24.Kaur, R., Pathania, R., Sharma, V., Mande, S. C., and Dikshit, K. L. (2002) Appl. Environ. Microbiol. 68 152-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner, P. R., Gardner, A. M., Martin, L. A., and Salzman, A. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 10378-10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner, P. R. (2008) Methods Enzymol. 436 213-233 [Google Scholar]
- 27.Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003) Nucleic Acids Res. 31 3381-3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., and Klein, M. L. (1983) J. Chem. Phys. 79 926-935 [Google Scholar]
- 29.Pearlman, D. A., Case, D. A., Caldwell, J. W., Ross, W. R., Cheatham, T. E., III, DeBolt, S., Ferguson, D., Seibel, G., and Kollman, P. (1995) Comput. Phys. Commun. 91 1-41 [Google Scholar]
- 30.Amadei, A., Linssen, A. B. M., and Berendsen, H. J. C. (2004). Proteins 17 412-425 [DOI] [PubMed] [Google Scholar]
- 31.Cohen, J., Arkhipov, A., Braun, R., and Schulten, K. (2006) Biophys. J. 91 1844-1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen, J., and Schulten, K. (2007) Biophys. J. 93 3591-3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, B. J., Cohen, J., Welford, R. W., Pearson, A. R., Schulten, K., Klinman, J. P., and Wilmot, C. M. (2007) J. Biol. Chem. 282 17767-17776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen, J., Olsen, K. W., and Schulten, K. (2008) Methods Enzymol. 437 437-455 [DOI] [PubMed] [Google Scholar]
- 35.Scott, E. E., Gibson, Q. H., and Olson, J. S. (2001) J. Biol. Chem. 276 5177-5188 [DOI] [PubMed] [Google Scholar]
- 36.Pawaria, S., Lama, A., Raje, M., and Dikshit, K. L. (2008) Appl. Environ. Microbiol. 74 3512-3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vuletich, D. A., and Lecomte, J. T. (2006) J. Mol. Evol. 62 196-210 [DOI] [PubMed] [Google Scholar]
- 38.Petr̂ek, M., Otyepka, M., Banás, P., Košinová, P., Koča, J., and Damborsky, J. (2006) BMC Bioinformatics 7 316-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey, W., Dalke, A., and Schulten, K. (1996) J. Mol. Graph. 14 33-38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.