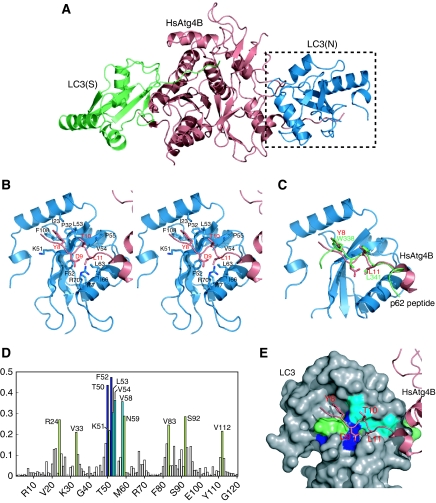
Figure 5.
Interaction of the N-terminal tail with the WXXL-binding site of LC3. (A) Overall structure of HsAtg4B bound to both substrate and non-substrate LC3. Substrate LC3 is designated as LC3(S) and coloured green, whereas non-substrate LC3 is designated as LC3(N) and coloured blue. (B) Stereo-view of the N-terminal tail of HsAtg4B bound to the WXXL-binding site of LC3. The side chains of the residues involved in the interaction between HsAtg4B and LC3 are labelled and shown with a stick model. Model colouring is the same as in (A). (C) Structural comparison of the N-terminal tail–LC3 interaction with the p62–LC3 interaction. The structure of LC3 complexed with a p62 peptide is superimposed on that complexed with the N-terminal tail peptide. As the structure of LC3 is almost identical to each other, only that bound to the N-terminal tail peptide of HsAtg4B is shown. (D) Chemical shift perturbations of the LC3 backbone amide groups upon complex formation with the N-terminal-tail peptide. The combined 1H and 15N chemical shift differences, calculated using the equation Δp.p.m.=[(ΔδHN)2+(ΔδN/5)2]1/2 were plotted against residue numbers. (E) Mapping of chemical shift perturbation results on the crystal structure of LC3 bound to HsAtg4B. The residues with Δp.p.m. >0.4 are shown in blue, 0.4>Δp.p.m.>0.3 in cyan and 0.3>Δp.p.m. >0.2 in green.