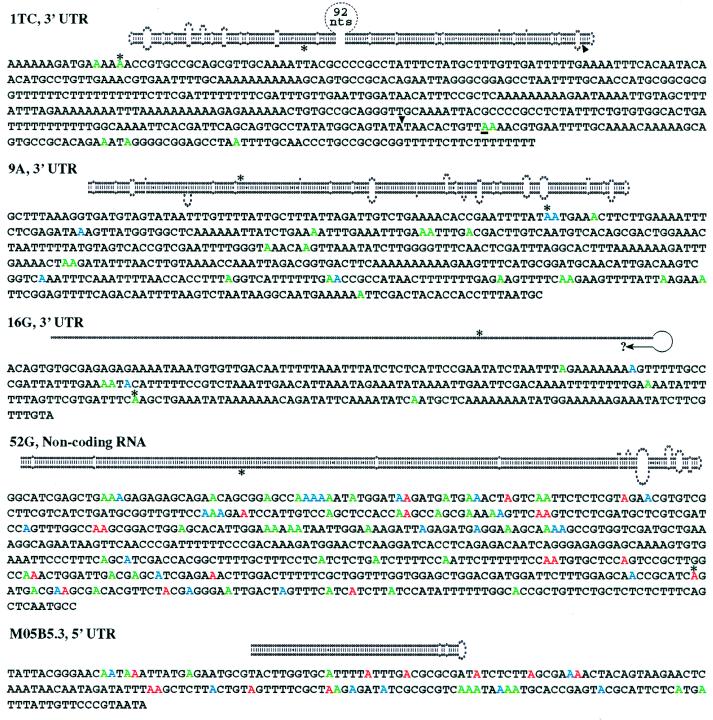
Figure 3.
The predicted structure for each ADAR substrate is shown above its sequence, with observed editing sites shown in color. Ten cDNAs were sequenced for 1TC and 52G and six for all others. Adenosines edited in >70% of the cDNAs are red, those in 40–70% blue, and less than 40% green. For each cDNA, the percentage of total adenosines that appeared as guanosines was determined. 1TC (0–3%), 9A (2–7%), and 16G (0–4%) were selectively edited, whereas 52G (10–22%) and M05B5.5 (23–32%) were deaminated more promiscuously. ∗, RNase T1 cleavage sites detected by differential display; arrowhead (1TC), Tc1 insertion site; question mark (16G), unfinished sequence not yet confirmed by our independent sequencing. The most efficiently edited A in 1TC is underlined. Conclusions about editing site locations in M05B5.3, 9A, and 16G are based on ORFs predicted by genefinder and on our cDNA sequencing. cDNA sequences revealed that two introns predicted by genefinder are not removed by splicing. These are the predicted first intron of M05B5.3 and the predicted last intron of ZC239.6 (9A). Although the edited region in the pop-1 (1TC) message was previously reported to lie within an intron (23), our PCR and Northern blot analyses indicate the region is in the 3′ UTR. cDNA sequences corresponding to 7339–8109 of F55A4 showed 7339 is an SL2 trans-splicing site and 7681–7225 is an intron. A likely polyadenylation signal begins at 8178 that would generate a transcript of a size consistent with Northern analyses. The number of nucleotides sequenced beyond the ends of the indicated structures were as follows (5′, 3′): 1TC (167, 203), 9A (370, 91), and MO5B5.3 (26, 74).