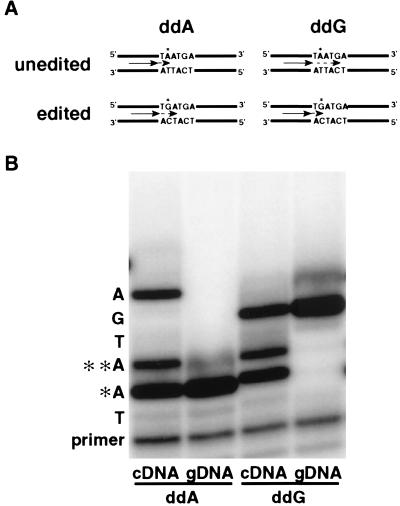
Figure 4.
Limited primer extension assay. (A) Principle: 5′ end-labeled primers (arrows) are extended in the presence of three deoxynucleotides and one dideoxynucleotide (see Methods). With ddA, primers annealed to unedited molecules are extended to the editing site (∗), whereas primers annealed to edited molecules continue to the next A (sense strand). With ddG, edited molecules show a new stop compared with unedited sequences. Dashed arrows indicate primer extension. (B) An autoradiogram showing primer extension products from 9A cDNA or genomic DNA (gDNA). Additional bands in the cDNA lanes verified that the RNase T1 cleavage site (∗) and the next adenosine (∗∗) were edited. Quantification indicated 48% of the 9A RNA molecules were edited at the T1 cleavage site (∗). In similar experiments we quantified the T1 cleavage site of 16G (5%) and the most efficiently edited site in 1TC (25%; underlined, Fig. 3). Assays of the T1 cleavage site in 52G gDNA and cDNA verified the editing site and showed it was edited ≥25%. However, the 52G structure made cDNA synthesis so inefficient that trace amounts of contaminating gDNA were coamplified to become a significant fraction of the sample; thus an exact number could not be obtained.