Abstract
Temporal coordination of hepatic drug-processing gene (DPG) expression facilitates absorption, biotransformation, and excretion of exogenous and endogenous compounds. To further elucidate the circadian rhythm of hepatic DPG expression, male C57BL/6 mice were subjected to a standard 12-h light/dark cycle, and livers were collected at 2:00, 6:00, and 10:00 AM and 2:00, 6:00, and 10:00 PM. The mRNAs of hepatic phase I enzymes (cytochromes P450, aldehyde dehydrogenases, and carboxylesterases), phase II enzymes (glucuronosyltransferases, sulfotransferases, and glutathione S-transferases), uptake and efflux transporters, and transcription factors were quantified. Messenger RNAs of various genes were graphed across time of day and compared by hierarchical clustering. In general, the mRNA of phase I enzymes increased during the dark phase, whereas the mRNAs of most phase II enzymes and transporters reached maximal levels during the light phase. The majority of hepatic transcription factors exhibited expression peaks either before or after the onset of the dark phase. During the same time period, the negative clock regulator gene Rev-Erbα and the hepatic clock-controlled gene Dbp also reached mRNA expression peaks. Considering their important role in xenobiotic metabolism, hepatic transcription factors, such as constitutive androstane receptor, pregnane X receptor, aryl hydrocarbon receptor, and peroxisomal proliferator activated receptor α, may be involved in coupling the hepatic circadian clock to environmental cues. Taken together, these data demonstrate that the circadian expression of the DPG battery and transcription factors contribute to the temporal detoxification cycle in the liver.
The mammalian circadian system temporally coordinates proliferation and metabolism in the body. Disruption of the circadian rhythm, as observed in rotating shift workers, has been reported to lead to an increased incidence of breast and colon cancers (Haus and Smolensky, 2006). Xenobiotic detoxification systems in liver, intestine, and kidney, among many other metabolic processes, are subjected to the hierarchical coordination by the central clock in the suprachiasmatic nucleus and the peripheral clock in each organ (Levi and Schibler, 2007). Optimizing drug dosing time has been suggested to be an important consideration for the treatment of several chronic diseases and cancer (Baraldo, 2008).
The liver is the major tissue for drug metabolism and contains most of the drug-processing genes (DPGs) in the body. Hepatic DPGs include genes encoding phase I and II metabolizing enzymes, as well as uptake and efflux transporters.
Hepatobiliary transporters play important roles in the uptake and clearance of xenobiotics by the liver, contributing to the “first-pass effect.” The hepatic solute carrier family of proteins mediate uptake of xenobiotics across the sinusoidal membrane. These transporters include Na+-taurocholate-cotransporting polypeptide, organic anion-transporting polypeptide (Oatp) 1a1, 1a4, 1b2, and 2b1, organic anion transporter 2 (Oat2), organic cation transporter 1 (Oct1), and equilibrative nucleoside transporter 1 (Klaassen and Lu, 2008). Xenobiotics or their metabolites are transported out of hepatocytes into bile by canalicular transporters including multidrug resistance-associated protein 2 (Mrp2), breast cancer resistance protein (Bcrp), multidrug resistance protein 1 (Mdr1), multi-drug and toxin extrusion 1 (Mate1), and the bile salt export pump. Another excretory route of liver is transporting xenobiotics and their metabolites back into the blood to be eliminated by kidneys into urine. Transporters Mrp3, 4, and 6, situated on the sinusoidal membrane of hepatocytes, are responsible for this “alternative efflux” process. On the canalicular membrane of hepatocytes and cholangiocytes, the flippase ATPase 8b1 (Atp8b1) translocates aminophospholipids from the outer to inner leaflet, whereas Mdr2 translocates phosphatidylcholine from the inner to outer leaflet.
Xenobiotic metabolism within hepatocytes consists of two major divisions. Phase I enzymes, including cytochromes P450 (P450), alcohol dehydrogenases, aldehyde dehydrogenases (Aldhs), carboxylesterases (Cess), epoxide hydrolases, NAD(P)H:quinone oxidoreductases (Nqos), and paraoxonases (Pons), carry out oxidation, reduction, and hydrolysis reactions on substrates to introduce or unmask functional groups (Parkinson and Ogilvie, 2008). Metabolites of phase I reactions are more water-soluble. In many cases, functional groups produced by phase I metabolism are further conjugated with glucuronic acid, sulfate, glutathione, or amino acids, catalyzed by phase II enzymes UDP-glucuronosyltransferases (Ugts), sulfotransferases (Sults), glutathione S-transferases (Gsts), and N-acetyltransferases, respectively. In general, conjugation reactions markedly increase the hydrophilicity of xenobiotics and enhance their excretion into bile and/or blood.
Hepatic transcription factors play crucial roles in regulating the expression of DPGs. Upon activation, transcription factors release corepressors and recruit coactivators and initiate target gene expression by binding to specific response elements within regulatory regions. Nuclear receptors, such as constitutive androstane receptor (CAR), pregnane X receptor (PXR), peroxisomal proliferator-activated receptor α (PPARα), hepatocyte nuclear factor 4α (HNF4α), and other transcription factors, including aryl hydrocarbon receptor (AhR), nuclear factor erythroid 2-related factor 2 (Nrf2), HNF1α, and CAAT/enhancer-binding proteins (C/EBPs), are key mediators in coordinating DPG expression. Moreover, CAR, PXR, and AhR are recognized as xenobiotic sensors. They regulate a battery of phase I and II enzymes, as well as membrane transporters to accelerate the elimination of toxicants (Klaassen and Slitt, 2005; Nakata et al., 2006).
Diurnal rhythms in catalyzing activity of rodent hepatic enzymes have been known for decades. For example, circadian rhythms were demonstrated in the metabolism of aminopyrine and p-nitroanisole in male rats and mice in the 1960s (Radzialowski and Bousquet, 1968). The profound effect of the circadian clock on drug metabolism was highlighted recently in mice with mutations in three circadian-controlled PARbZip transcription factors, including albumin D site-binding protein (Dbp). These triple mutant mice have increased sensitivity to xenobiotics and premature aging (Gachon et al., 2006). Thus, the circadian oscillator, consisting of the core circadian transcription factors Clock, Bmal1, Rev-Erbα, Periods, and Cryptochromes, together with circadian-controlled genes, such as the PARbZip transcription factors, adds another dimension to xenobiotic metabolism. However, less is known about the overall circadian expression of hepatic DPGs and the underlying regulatory mechanisms. Thus, in the present study we characterized the daily expression patterns of phase I and II drug-metabolizing enzymes, uptake and efflux transporters, hepatic transcription factors, and representative circadian transcription factors in mouse liver in an attempt to reveal their potential inter-relationships.
Materials and Methods
Animals. Animal experiments were performed in accordance with the institutional animal care and use committee guidelines. Male C57BL/6 mice (7 weeks of age) were purchased from Charles River Laboratories, Inc. (Wilmington, MA) and acclimated for 2 weeks in a temperature-humidity controlled facility with a standard 12-h light schedule (lights on at 5:00 AM and off at 5:00 PM). All animals were given ad libitum access to water and standard rodent chow (Harlan Teklad 8604; Harlan Teklad, Madison, WI). Mice (n = 5/time point) were anesthetized within 20 min before 2:00, 6:00, and 10:00 AM and 2:00, 6:00, and 10:00 PM. Livers were collected, rinsed with ice-cold saline, frozen in liquid nitrogen, and stored at –80°C.
Multiplex Suspension Array. Total RNA was extracted from livers using RNA-Bee reagent (Tel-Test Inc., Friendswood, TX) according to the manufacturer's protocol and was quantified spectrophotometrically. Equal amounts of RNA from individual samples were combined to make a pooled sample for each time point. The mRNA expression of genes of interest in livers was determined by Panomics 1.0/2.0 QuantiGene plex technology (Panomics Inc., Fremont, CA). Panels were designed by Panomics, and individual gene information can be found by searching for the following plex set numbers on the Panomics Web site (http://www.panomics.com): 20021, 2051, 2053, 2055, 2058, and 2061. In brief, in a 96-well plate, 10 μg of pooled RNA sample (n = 5/time point) was added to wells containing X-MAP beads coated with capture probes, label extenders, and blockers. The plate was incubated overnight at 54°C. Beads and bound target mRNA were then transferred to filter plates, washed three times, and incubated with amplifier at 46°C for 1 h. Wells were then washed twice and incubated with label (biotin) at 46°C for 1 h, washed again twice, and incubated with streptavidin-conjugated R-phycoerythrin, which binds biotinylated probes, and incubated at room temperature for 30 min. Streptavidin-conjugated R-phycoerythrin fluorescence was analyzed using a Bio-Plex 200 system array reader with Luminex 100 X-MAP technology, and data were acquired using Bio-Plex data manager software 4.1 (Bio-Rad, Hercules, CA). mRNA expression is presented as relative light units per 10 μg of total RNA. In all graphs of this study, peak stands for the highest gene expression level and trough is the lowest during a 24-h period.
Branched DNA Assay. To verify the circadian expression patterns of genes displaying strong or weak signals in multiplex suspension assay, a branched DNA (bDNA) signal amplification assay (Quantigene High Volume bDNA Signal Amplification Kit; Panomics Inc.) was performed for individual genes. For highly expressed genes (Cyp2e1, Cyp3a11, Ces5b1, and Pon1), 1 μg of total RNA was added per well. For lowly expressed genes [Cyp2b10, Mdr1, Kelch-like ECH-associated protein 1 (Keap1), and AhR], 10 μg of RNA was loaded per well. Probe sets for these genes were published previously (Cheng et al., 2005b; Cheng and Klaassen, 2006; Petrick and Klaassen, 2007), and the bDNA procedure was performed as described previously (Hartley and Klaassen, 2000). In brief, pooled RNA was added to each well of a 96-well plate containing premixed capture hybridization buffer (50 μl) and diluted probe set (50 μl). Overnight hybridization between RNA and probe sets was performed at 53°C. Subsequent hybridization and posthybridization wash steps were carried out according to the manufacturer's protocol, and luminescence was quantified with a Synergy 2 Multi-Detection Microplate Reader interfaced with Gen5 Reader Control and Data Analysis software (Bio-Tek Instruments, Winoosky, VT).
Expression Profiles by Hierarchical Clustering. For each gene, mRNA expression at each time point (2:00, 6:00, and 10:00 AM and 2:00, 6:00, and 10:00 PM) was expressed as the ratio of expression at 6:00 AM. Patterns of gene expression across time were distinguished by hierarchical clustering (JMP 7; SAS Institute, Cary, NC). Peak expression for each gene is represented by the darkest region of the gray scale spectrum, with the lowest expression indicated by the lightest (gray) region. Each spectrum is specific to the scale of mRNA expression for each gene on an individual basis. Therefore, the relative intensities of gray scale spectra cannot be compared among genes.
Results
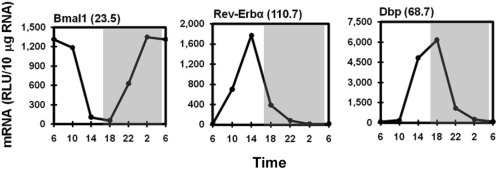
Diurnal Expression of Circadian Transcription Factors in the Liver. As expected, components of the circadian clock displayed the most marked oscillations (Fig. 1). mRNA of the core circadian transcription factor, Bmal1, was robust between 2:00 and 10:00 AM and only approximately 4.3% of the maximal level between 2:00 and 6:00 PM. Rev-Erbα, the important negative clock regulator, oscillated in antiphase to Bmal1. Rev-Erbα mRNA at 2:00 PM was approximately 110-fold higher than at 10:00 PM and 6:00 AM. The important hepatic clock-controlled gene Dbp, another liver-enriched transcription factor, was also expressed in antiphase to Bmal1. Similar to that of Rev-Erbα, the mRNA of Dbp was markedly increased at 2:00 PM and by 6:00 PM had reached a peak that was 68.7-fold higher than at 2:00 and 6:00 AM.
Fig. 1.
Rhythmic transcripts of prototypical circadian oscillators (Bmal1 and Rev-Erbα) and clock-controlled gene (Dbp) in mouse liver. Total RNA from mouse livers (n = 5 per time point) was pooled, and mRNA expression was determined by a multiplex suspension array. Data at 6:00 AM were used twice to graph a 24-h cycle of mRNA expression. Numbers in parentheses next to the gene names indicate the peak/trough ratio of each mRNA. The shaded area of the figure represents the dark phase, and the white area represents the light phase. The same design is also used in the following figures. RLU, relative light units.
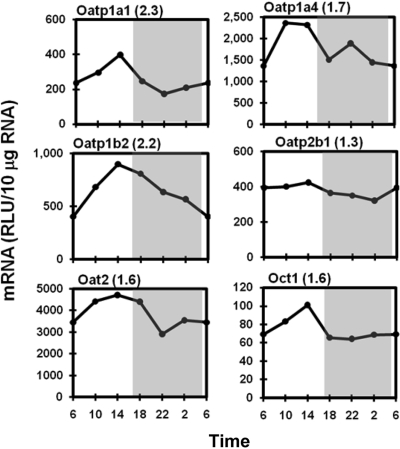
Diurnal Expression Profiles of Hepatic Uptake Transporters. The transcripts of Oatps tended to be more abundant during the light phase than during the dark phase. As shown in Fig. 2, liver-enriched Oatp1a1 and Oatp1a4 and the liver-specific Oatp1b2 (Cheng et al., 2005a) uniformly peaked at approximately 2:00 PM with the peak/trough ratio varying from 1.7 to 2.3. Oatp2b1 mRNA had a minimal fluctuation over 24-h in mouse livers. The mRNA of Oat2 was higher between 10:00 AM and 6:00 PM, with a 1.6-fold peak/trough ratio. Similar to hepatic Oatps, the electrogenic uniporter Oct1 exhibited a single 60% increase of mRNA at 2:00 PM and remained consistently low during the dark phase. No significant daily fluctuations were observed in expression of the bile acid transporter Na+-taurocholate-cotransporting polypeptide or the bidirectional nucleoside transporter equilibrative nucleoside transporter 1 (data not shown).
Fig. 2.
Daily mRNA expression profiles of basolateral uptake transporters in mouse liver. Graphic presentation follows that outlined in the legend to Fig. 1. RLU, relative light units.
Daily Variation in the Expression of Phase I Enzymes. The mRNAs of major microsomal P450s tended to accumulate more in the dark phase. As shown in Fig. 3, Cyp2b10 mRNA displayed a robust circadian rhythm with a peak/trough ratio of 2.8; the highest expression occurred at 2:00 AM and the lowest at 2:00 PM. Cyp2e1 and Cyp3a11 demonstrated the highest mRNA levels at 10:00 PM and 2:00 AM, with peak/trough ratios of 1.6 and 1.8, respectively. Cyp4a14 mRNA exhibited marked fluctuation with a 5-fold peak/trough ratio during the dark phase. Whereas Cyp4a14 expression was relatively consistent during the light phase, its mRNA reached the highest expression at 6:00 PM, then rapidly declined to the lowest level in the middle of the dark phase (10:00 PM–2:00 AM), and returned to a higher level in the light phase. The daily change in Cyp1a2 mRNA is 1.3-fold, with the peak at 6:00 PM (data not shown). Cytochrome P450 reductase (Cpr), the common electron donor for P450s, presented a marked circadian fluctuation of mRNA with a 3.8-fold peak/trough ratio. Cpr mRNA was robustly expressed at 6:00 and 10:00 PM, and remained at a low level during the day. The active site of cytochrome P450 contains a heme iron center. δ-Aminolevulinic acid synthase (Alas1) is the rate-limiting enzyme in the synthesis of heme. Messenger RNA of Alas1 oscillated with a 7.8-fold magnitude and peaked at 6:00 PM.
Fig. 3.
Daily mRNA expression profiles of major cytochrome P450 enzymes in mouse liver. Graphic presentation follows that outlined in the legend to Fig. 1. *, branched DNA assay was used to determine the mRNA expression of Cyp2e1 and Cyp3a11 with 1 μg of pooled RNA (n = 5 per time point) and 10 μg for Cyp2b10. RLU, relative light units.
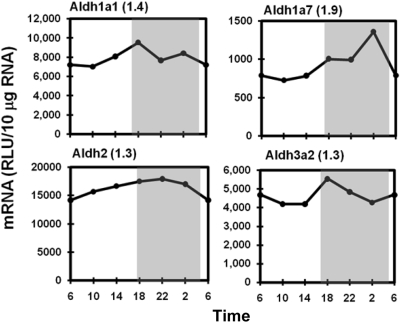
The Aldh enzyme family carries out a diversity of metabolic functions, including detoxification. This study quantified mRNA of Aldh1a1, 1b1, 2, 3a2, 4a1, 6a1, 7a1, 8a1, and 9a1 in mouse liver. In general, the Aldh mRNAs tended to be higher in the dark phase, but the variations were less than 1.5-fold. The circadian expression patterns of Aldh1a1, 1a7, 2, and 3a2 are shown in Fig. 4 as representatives, whereas the other Aldhs are not shown. Aldh1a1 and Aldh3a2 peaked at 6:00 PM. Aldh1a7 displayed the highest fluctuation among the Aldhs, with a peak (peak/trough ratio 1.9) at 2:00 AM. Aldh2 mRNA was expressed equally during the 24-h cycle.
Fig. 4.
Daily mRNA expression profiles of representative Aldhs in mouse liver. Graphic presentation follows that outlined in the legend to Fig. 1. RLU, relative light units.
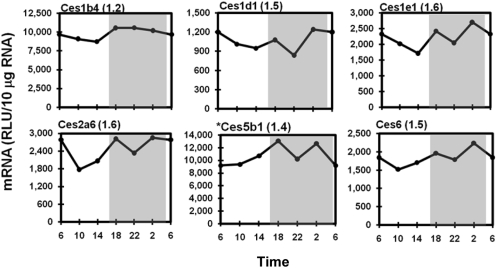
Hepatic Ces isoenzymes hydrolyze ester- and amide-containing compounds. Figure 5 illustrates typical circadian variations of Ces mRNAs, namely Ces1b4 (also known as Esd), 1d1, 1e1, 2a6, 5b1, and 6. Expression profiles of Ces1g2, 1h1, and 3a2 are not shown because of small daily variations; peak/trough values were 1.1, 1.2, and 1.4, respectively. Similar to the Aldhs, the majority of the Ces transcripts displayed mild fluctuations in mRNA over time, with slightly higher levels during the dark. The peak/trough ratio ranged from 1.1 (Ces1g2 and Ces5b1) to 1.6 (Ces1e1 and Ces2a6). It is interesting to note that many Ces mRNAs, including Ces1d1, 1e1, 2a6, 5b1, and 6, exhibited an additional minor trough in the middle of the dark phase (10:00 PM).
Fig. 5.
Daily mRNA expression profiles of representative Cess in mouse liver. Graphic presentation follows that outlined in the legend to Fig. 1. *, expression profile of Ces5b1 was quantified by bDNA with 1 μg of pooled RNA (n = 5/time point). RLU, relative light units.
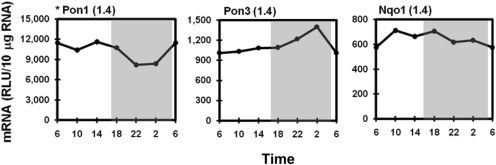
Pons hydrolyze a broad range of organophosphorus esters. Pon1 and Pon3 are the two isoforms highly expressed in mouse liver. Both Pons were evenly expressed during the light phase. However, their expression profiles were divergent during the dark phase. Pon1 exhibited an approximately 40% decrease in mRNA at 10:00 PM and 2:00 AM, whereas Pon3 mRNA increased 40% at 2:00 AM (Fig. 6). Figure 6 also shows the flat daily expression pattern of the cytosolic flavoprotein Nqo1, which catalyzes the reduction of quinones and protects hepatocytes against oxidative stress.
Fig. 6.
Daily mRNA expression profiles of paraoxonases (Pons) and NAD(P)H-quinone oxidoreductase 1 (Nqo1) in mouse liver. Graphic presentation follows that outlined in Fig. 1. *, expression profile of Pon1 was quantified by bDNA with 1 μgofRNA from 5 mouse livers per time point. RLU, relative light units.
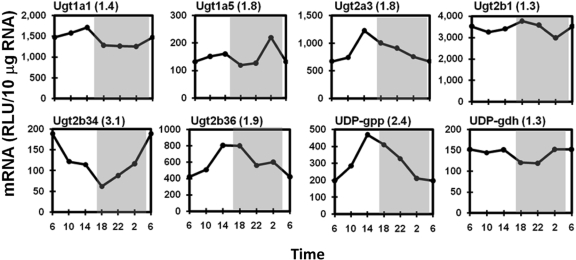
Daily Variation in the Expression of Phase II Enzymes. The circadian expression patterns of major hepatic UGTs are shown in Fig. 7. Ugt1a1, 1a6, and 1a9 exhibited slightly higher mRNA expression in the late-light phase (peak/trough values between 1.3 and 1.5; represented by Ugt1a1 in Fig. 7), whereas Ugt1a5 is different from the other Ugt1a mRNAs by displaying a brief 1.8-fold mRNA peak at 2:00 AM. The liver-specific Ugt2b1 exhibited slightly higher (30%) mRNA expression around the light/dark transitions (6:00 AM and 6:00 PM). Similar to most of the Ugt1a mRNAs, several Ugt2 family members also exhibited the highest mRNA expression in the late-light phase (2:00 PM), including Ugt2a3 (1.8-fold), Ugt2b35 (1.6-fold, not shown), and Ugt2b36 (1.9-fold). In contrast to other Ugts, Ugt2b34 displayed a unique circadian pattern, with the highest mRNA level at 6:00 AM and the lowest at 6:00 PM. In mouse liver, Ugt3a1 and Ugt3a2 expression was slightly higher in the light phase (peak/trough values of 1.4 and 1.3, respectively; data not shown). The two enzymes involved in the biosynthesis of UDP-glucuronic acid (UDP-GA), the cosubstrate for glucuronidation, exhibited different circadian expression patterns. The mRNA of UDPG-pyrophosphorylase, the enzyme that catalyzes the formation of UDP-glucose from glucose 1-phosphate and UTP, exhibited abundant circadian rhythmicity, with a 2.4-fold peak at 2:00 PM. However, UDPG-dehydrogenase, the enzyme that subsequently oxidizes UDP-glucose to UDP-GA, exhibited only a 1.3-fold variation in mRNA.
Fig. 7.
Representative circadian expression profiles of hepatic Ugt isoforms and UDP-GA biosynthesis enzymes. Graphic presentation follows that outlined in the legend to Fig. 1. RLU, relative light units.
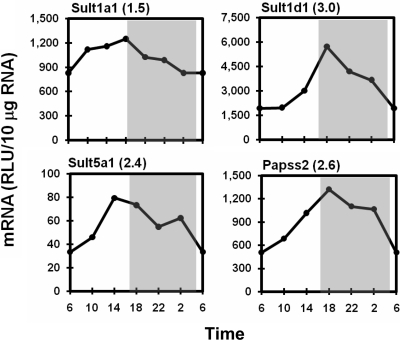
Among the seven hepatic Sult isoforms quantified in livers of male mice, Sult1a1 and Sult1d1 exhibited high-level mRNA expression, mRNA expression for Sult5a1 was moderate, whereas mRNAs of Sult1c1, 1e1, 2a2, and 3a1 were not detected. mRNA levels of Sult1a1, 1d1, and 5a1 oscillated with 1.5-, 3.0-, and 2.4-fold amplitudes, respectively, with peaks appearing around the day to night transition (Fig. 8). 3′-Phosphoadenosine-5′-phosphosulfate (PAPS) provides sulfonate groups for sulfoconjugation catalyzed by all Sults. PAPS synthase 2 (Papss2) is the rate-limiting enzyme for the production of PAPS in liver. Papss2 mRNA reached peak levels at 6:00 PM and remained elevated until 2:00 AM and then decreased to a trough at 6:00 AM (peak/trough ratio 2.6).
Fig. 8.
Representative rhythmic expression patterns of hepatic Sult isoforms and PAPS synthase. Graphic presentation follows that outlined in the legend to Fig. 1. RLU, relative light units.
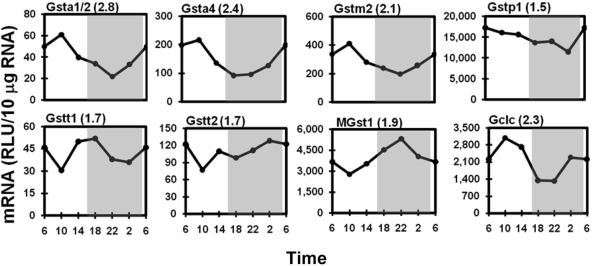
Many isoforms of Gsts are abundantly expressed in liver. Gsta1/2 (2.8-fold) and Gsta4 (2.4-fold) transcripts exhibited the largest daily fluctuations (Fig. 9), both displaying the highest expression in the early light phase (10:00 AM) and lowest in the middle-dark phase (at approximately 10:00 PM). Circadian rhythms of the five mu family Gsts are similar to those of the two alpha Gsts. Gstm2, exhibiting a 2.1-fold daily fluctuation in mRNA expression, is shown in Fig. 9, whereas circadian variations of Gstm1, m3, m4, and m6, with peak/trough ratios from 1.3 to 1.6, are not shown. Gstp1 was the most abundantly expressed Gst isoform, which had slightly higher (1.5-fold peak/trough value) mRNA accumulation in the light phase than in the dark phase. Different from the light-phase predominant pattern described above, the lowest expression of the theta class of Gsts, Gstt1 and Gstt2, was at 10:00 PM. Gstt1 had higher expression at both of the light-dark transition time points (6:00 AM and 6:00 PM), and Gstt2 mRNA was expressed more during the dark phase. Peak/trough ratios were 1.7-fold for both Gstts. The abundantly expressed mRNA of the microsomal Gst isoform MGst1 oscillated with a 1.9-fold magnitude, with peak and trough appearing at 10:00 PM and 10:00 AM, respectively. The catalytic subunit of glutamate-cysteine ligase (Gclc) is the rate-limiting enzyme for glutathione synthesis. Gclc expression exhibited circadian variation with the peak appearing at 10:00 AM and trough between 6:00 and 10:00 PM, with a peak/trough ratio of 2.3.
Fig. 9.
Representative circadian expression profiles of hepatic Gst isoforms and Gclc. Graphic presentation follows that outlined in the legend to Fig. 1. RLU, relative light units.
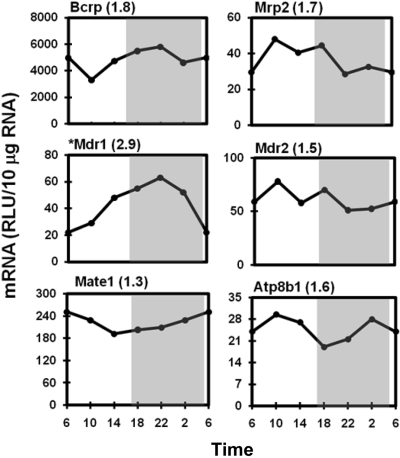
Diurnal Expression of Hepatic Efflux Transporters. The mRNA expression of major drug transporters on the canalicular membrane of hepatocytes oscillated less than 2-fold (Fig. 10), including the efflux transporters Bcrp (1.8-fold), Mrp2 (1.7-fold), Mdr2 (1.5-fold), and Mate1 (1.3-fold), as well as the aminophospholipid flippase ATPase 8b1 (1.6-fold). However, Mdr1 is the exception that exhibited a 2.9-fold fluctuation, with maximal expression at 10:00 PM. The expression of transporters on the sinusoidal membrane of hepatocytes, namely Mrp3, Mrp4, and Mrp6, did not exhibit diurnal fluctuations (peak/trough values between 1.2- and 1.5-fold; data not shown).
Fig. 10.
Daily mRNA expression profiles of canalicular transporters in mouse liver. Graph presentation follows that outlined in the legend to Fig. 1. *, expression of Mdr1 mRNA was quantified by bDNA assay with 10 μg of pooled RNA (n = 5/time point). RLU, relative light units.
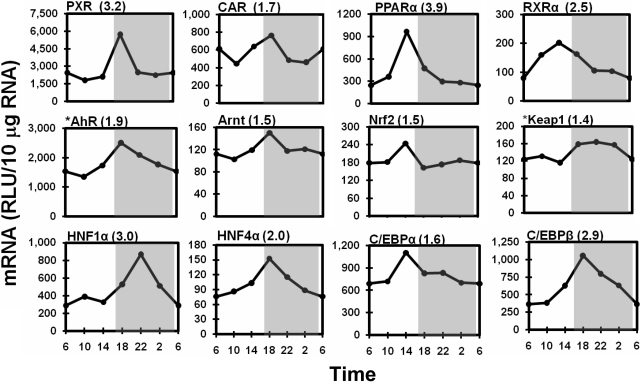
Temporal Expression Profiles of Hepatic Transcription Factors. The majority of hepatic transcription factors examined exhibited peak mRNA expression around the light/dark transition (2:00–10:00 PM) (Fig. 11). The xenobiotic sensors, PXR and CAR, both peaked at 6:00 PM, with 3.2- and 1.7-fold amplitudes, respectively. PPARα and the common nuclear receptor heterodimer partner retinoid X receptor α reached 3.9- and 2.5-fold peaks at 2:00 PM, respectively. Similar to PXR and CAR, AhR and its nuclear heterodimer partner, aryl hydrocarbon receptor nuclear translocator, both demonstrated peak mRNA levels at 6:00 PM, with 1.9- and 1.4-fold peak/trough ratios, respectively. Nrf2 expression was consistent across the 24-h time course, except for a 50% increase at 2:00 PM, whereas the expression of its cytoplasmic repressor Keap1 was higher (40%) during the dark phase than during the light phase.
Fig. 11.
Diurnal expression profiles of important xenobiotic-metabolism transcription factors in the liver. Graphic presentation follows that outlined in the legend to Fig. 1. *, messenger RNA expression of Keap1 and AhR was quantified by bDNA assay with 10 μg of pooled RNA (n = 5/time point). RLU, relative light units.
Furthermore, expression of several liver-enriched transcription factors also exhibited dramatic circadian rhythms in mouse liver (Fig. 11). HNF1α mRNA expression did not increase until the onset of the dark phase, whereby a 3-fold peak expression was reached at 10:00 PM. HNF4α mRNA increased to the maximum at 6:00 PM, with a 2-fold peak/trough ratio. C/EBPα expression exhibited a 1.6-fold peak before the onset of the dark phase at 2:00 PM, whereas C/EBPβ mRNA steadily increased to a 2.9-fold peak at 6:00 PM and then gradually decreased during the dark phase.
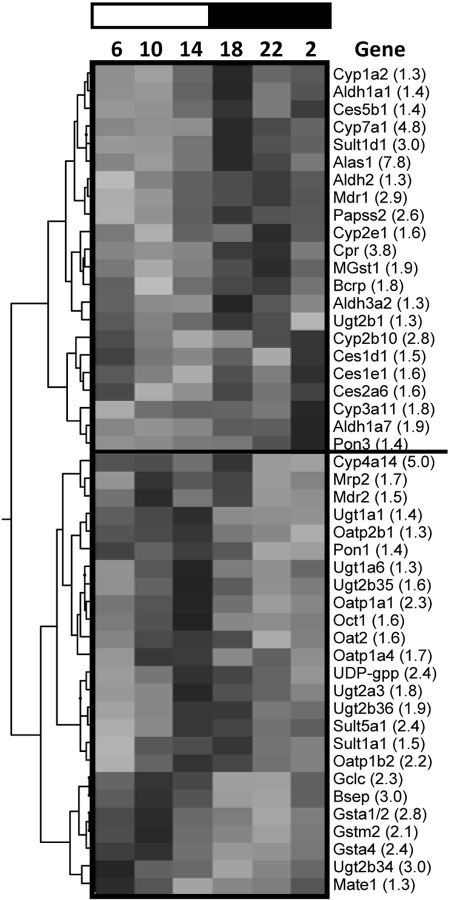
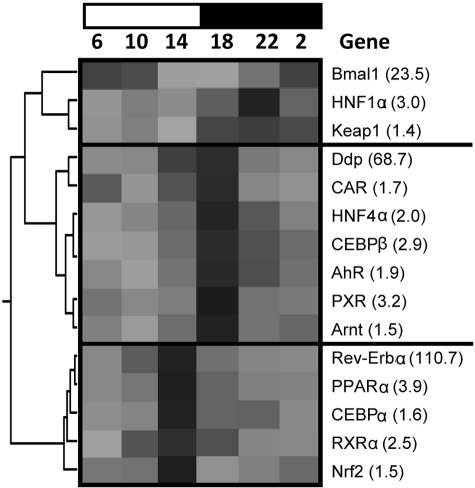
Hierarchical Cluster Analysis of the Circadian Expression Profiles of Hepatic DPGs and Transcription Factors. The expression profiles of each DPG and transcription factor across time were analyzed by hierarchical cluster analysis, whereby peak mRNA expression (by time) corresponds with the darkest shaded region of the heat map (Fig. 12). The circadian expression of hepatic DPGs can be broadly classified into dark-peak or light-peak groups. It is interesting to note that genes exhibiting peaks during the dark phase (6:00 and 10:00 PM and 2:00 AM) include almost all phase I enzymes but only a few phase II enzymes (e.g., Sult1d1, Papss2, MGst1, and Ugt2b1) and transporters (e.g., Mdr1 and Bcrp). In contrast, the majority of phase II conjugation enzymes and transporters exhibit the highest mRNA expression in the light phase (6:00 and 10:00 AM and 2:00 PM). Transcription factors can be classified into three major clusters (Fig. 13): the 10:00 PM peak cluster, led by the core clock component Bmal1, comprises transcription factors HNF1α and Keap1; the 2:00 PM peak cluster, led by Rev-Erbα, includes PPARα, Nrf2, C/EBPα, and retinoid X receptor α; and the 6:00 PM peak cluster, led by Dbp, consists of CAR, PXR, HNF4α, AhR, Arnt, and C/EBPβ.
Fig. 12.
Hierarchical cluster analysis of temporal expression profiles of major drug-processing genes. Each column represents a circadian time point, and each row represents a gene with the name indicated. The number in parentheses beside each gene name represents the mRNA expression peak/trough ratio of that particular gene. Numbers at the top depict the time of day, and the bar represents the light phase (white) or dark phase (black). Peak expression for each gene is represented by the darkest region of the gray scale spectrum and lowest expression indicated by the lightest (gray) region. Each spectrum is specific to the scale of mRNA expression for each gene on an individual basis. Therefore, comparing gene expression profiles across gray scale spectra is not valid because of differences in the relative gene expression.
Fig. 13.
Hierarchical cluster analysis of the diurnal expression of hepatic transcription factors. Graphic presentation follows that outlined in the legend to Fig. 12.
Discussion
Circadian variations in the absorption, distribution, metabolism, and excretion of drugs may influence drug efficacy in the clinical treatment of cancer, cardiovascular disease, bacterial infection, inflammation, and so on (Baraldo, 2008). Comprehensive analysis of circadian expression of hepatic DPGs in this report demonstrates a systemic and progressive temporal detoxification network in the liver.
The present study presents time-dependent variations in mRNA expression of phase I enzymes participating in reduction (Nqo1), hydrolysis (Cess and Pons), and oxidation (P450s and Aldhs) of xenobiotics. Higher P450 enzyme activities during the dark phase was previously observed in mouse livers (Radzialowski and Bousquet, 1968). Accordingly, mRNAs of several critical P450 enzymes are more highly expressed at night (Fig. 3). Previous reports on the circadian rhythms of Cyp2b10 (Gachon et al., 2006), Cyp2e1 (Matsunaga et al., 2008), and Cyp3a11 (Takiguchi et al., 2007) in mice are comparable to those in this study. In the dark phase, mouse liver robustly expresses Cpr and Alas1 to correspondingly supply Cyps with electrons and heme, respectively. On the basis of these gene expression profiles, higher P450-mediated drug-metabolizing ability is expected in the dark phase in the livers of mice.
Temporal mRNA expression profiles of phase II enzymes in this study support the rhythmic conjugation reactions observed previously in rodents. Glucuronidation is important for xenobiotic biotransformation (Tukey and Strassburg, 2000). Messenger RNA levels of the hepatic Ugts (e.g., Ugt1a1, 1a6, 1a9, 2a3, 2b35, and 2b36) are robust in the late-light phase (Figs. 7 and 12), suggesting that Ugt enzymes are increased during fasting. Expression of UDP-pyrophosphorylase oscillates in phase with the expression of Ugt enzymes to ensure the availability of the cosubstrate UDP-GA. The hepatic concentration of UDP-GA is highest during the fasting period and lowest during the feeding period (Howell and Klaassen, 1991). Likewise, reduced glutathione, the cosubstrate for glutathione conjugation, also exhibits a circadian rhythm in liver, with maximum concentration at 10:00 AM and minimal concentration at 6:00 PM (Schnell et al., 1984). Gclc and many Gsts (e.g., Gsta1/2, 1a4, m2, and p1) are maximally expressed during the early light phase (Figs. 9 and 12). Sulfation in mice is limited by sulfotransferase activity (Liu and Klaassen, 1996). The current study demonstrates that the male predominant Sults (Sult1a1, 1d1, and 5a1) (Alnouti and Klaassen, 2006) and the PAPS biosynthesis enzyme Papss2 have the highest mRNA expression during the light-to-dark transition. Taken together, phase II conjugation of xenobiotics in liver may have a circadian rhythm, with more glutathione conjugation in the early light phase, glucuronidation in the late-light phase, and sulfation in the early dark phase.
The circadian expression pattern of hepatic enzymes explains at least in part why the hepatotoxicity of xenobiotics, such as acetaminophen, exhibits a circadian rhythm. Kim and Lee (1998) showed that a single dose of acetaminophen at 8:00 PM produced marked hepatotoxicity in mice but almost no hepatotoxicity when given at 8:00 AM or 2:00 PM. Glucuronidation or sulfation enzyme activities in liver and concentrations of acetaminophen conjugates in plasma are similar when acetaminophen was administrated at the three times. However, higher glutathione concentrations were observed at 8:00 AM and lower P450 enzyme activity occurred at 2:00 PM. Therefore, it seems that lower biotransformation by P450s and higher hepatic glutathione levels, rather than Ugt- or Sult-mediated conjugation, protects livers from acetaminophen-induced liver injury. This speculation is further supported by the current study, in which higher expression of P450s was observed at night (Fig. 3), which corresponds to more hepatotoxicity produced by acetaminophen at 8:00 PM, whereas high expression of Gclc in the morning leads to higher hepatic glutathione levels, which provides protection from acetaminophen hepatotoxicity at 8:00 AM (Fig. 9).
Hepatic membrane transporters are involved in the absorption, distribution, and elimination processes of drugs and xenobiotics. However, the circadian rhythms of transporters are not well studied. The mRNAs of sinusoidal uptake transporters, represented by Oatps, coordinately increase before the day-to-night transition (Fig. 2), which suggests an anticipation of active hepatic absorption from the portal blood. Via hydrolysis of ATP, the ATP-binding cassette family of transporters actively excrete chemicals from the hepatocytes into bile or blood. Marked circadian rhythm is observed in the mRNA expression of canalicular transporters Mdr1 and bile salt export pump (data not shown), but fewer oscillations in the mRNA of other canalicular or sinusoidal efflux transporters (Fig. 10). Diurnal variations of Mdr1, Mdr2, and Mrp2 in the present study are consistent with findings in a previous report (Ando et al., 2005). However, the Mdr2 mRNA circadian profile differs from that found in another study (Kotaka et al., 2008). Rhythmic variations in the basal expression of membrane transporters may be important for the diurnal transport of nutrients, metabolic substances, or food-derived toxins in hepatocytes.
Collaboration of enzymes and transporters is an important consideration for hepatic detoxification processes. For instance, in human hepatocytes, the products oxidized by P450s and the products hydrolyzed by Cess can be further conjugated by UGTs colocalized in the endoplasmic reticulum lumen; the organic anion products by Ces-catalyzed hydrolysis and phase II conjugates are substrates for membrane transporters such as MRP2 or BCRP (Hosokawa et al., 2007). DPGs involved in a certain pathway can be simultaneously or sequentially expressed. As demonstrated in hierarchical analysis (Fig. 12), the mRNA expression of phase I enzymes is abundant in the dark phase, whereas the majority of phase II enzymes and transporters tend to be robustly expressed toward the end of the light phase. However, the highest expression of most DPGs is seen at 6:00 PM, the time to start feeding in mice. Taken together, the rhythmic expression of hepatic DPGs may underlie the time-dependent detoxifying capability of the liver.
Hierarchical analysis also reveals that hepatic transcription factors are expressed in-phase with both clock components and DPGs (Figs. 12 and 13). Thus, the hepatic transcription factors may couple the circadian clock and the daily detoxifying function of the liver. For example, the circadian rhythm of cholesterol 7α-hydroxylase is regulated not only by circadian transcription factors but also by common nuclear receptors such as liver X receptor α, PPARα, and HNF4α (Noshiro et al., 2007). Furthermore, the expression of nuclear receptors might be subjected to the regulation of the circadian clock. For instance, the circadian expression of CAR and its target gene Cyp2b10 in mouse liver is determined by PARbZip transcription factors (Gachon et al., 2006). Moreover, in rat liver, the phenobarbital-dependent induction of CYP2B1/2 mRNA displays diurnal differences attributable to the circadian expression of its major regulator CAR (Kanno et al., 2004).
The current study shows that several liver-enriched transcription factors, namely HNF1α, HNF4α, C/EBPα, and C/EBPβ, have clear circadian mRNA patterns. The circadian rhythms of these transcription factors are noteworthy because they are thought to regulate the expression of a number of DPGs. For example, mRNA expression of many enzymes and transporters is markedly reduced (Cyp1a2, Cyp2e1, Oat2, Oatp1a1, and Oatp1b2) or increased (Cyp4a14, Cyp7a1, Oct2, Mrp4, Mdr1a, and Mdr2) in HNF1α-null mouse livers (Maher et al., 2006).
It is noteworthy that the circadian clock can directly regulate DPG expression. In a recent study, the knockout of clock-controlled transcription factors including Dbp changed the expression of many hepatic DPGs involved in phase I (Cyp2b, Cyp2c, Ces3, and Cpr), phase II (Gstt1 and Gsta3), and transport (Abcg2) processes (Gachon et al., 2006). Taken together, xenobiotic-responsive and circadian transcription factors may together regulate the circadian expression of hepatic DPGs in response to fasting-to-feeding or light-to-dark transitions.
Xenobiotic-activated transcription factors can also affect the clock system. For example, the AhR ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin, is able to alter the expression of Per1 and Bmal1 and decrease phase shifts in response to light (Mukai et al., 2008). In addition, the mRNAs of AhR and its target gene Cyp1a1 both exhibit 2-fold diurnal fluctuations (Mukai et al., 2008). Another example is PPARα, which is not only regulated by CLOCK (Oishi et al., 2005) but also regulates Bmal1 expression in vivo via direct binding to the Bmal1 promoter (Canaple et al., 2006). Taken together, xenobiotic-activated transcription factors may also exert exogenous influences on the expression of clock components.
In summary, the current study describes the diurnal expression patterns of important DPGs and transcription factors in mouse liver. Phase I enzyme mRNA expression is highest during the dark phase, whereas phase II enzyme and transporter expression is abundant at different times throughout the light phase, thus underlying the temporal variation of drug-processing ability of the liver. The in-phase expression of hepatic transcription factors with most of the DPGs and several critical clock components suggests that transcription factors are important in coupling the circadian clock and DPG expression. The comprehensive characterization of diurnal patterns of important DPGs and transcription factors provides valuable references for drug development and clinical drug administration.
Acknowledgments
We thank Scott Reisman and Dr. Cheryl Rockwell for assistance with the Multiplex Suspension Array and numerous graduate students and postdoctoral fellows who helped to remove tissues from the mice.
This study was supported by the National Institutes of Health [Grants ES-09649, ES-09716, ES-07079, ES-018714, and RR-021940].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.024174.
ABBREVIATIONS: DPG, drug-processing gene; Oatp, organic anion-transporting polypeptide; Oat, organic anion transporter; Oct, organic cation transporter; Mrp/MRP, multidrug resistance-associated protein; Bcrp/BCRP, breast cancer resistance protein; Mdr, multidrug resistance; Mate 1, multidrug and toxin extrusion 1; Atp8b1, ATPase, class I, type 8b, member 1; P450, cytochrome P450; Aldh, aldehyde dehydrogenase; Ces, carboxylesterase; Nqo, NAD(P)H:quinone oxidoreductase; Pon, paraoxonase; Ugt/UGT, UDP-glucuronosyltransferase; Sult, sulfotransferase; Gst, glutathione S-transferase; CAR, constitutive androstane receptor; PXR, pregnane X receptor; PPARα, peroxisomal proliferator-activated receptor alpha; HNF, hepatocyte nuclear factor; AhR, aryl hydrocarbon receptor; Nrf2, nuclear factor erythroid 2-related factor 2; C/EBP, CAAT/enhancer-binding protein; Dbp, albumin D site binding protein; Bmal1, brain and muscle aryl hydrocarbon receptor nuclear translocator-like protein 1; bDNA, branched DNA; Keap1, Kelch-like ECH-associated protein 1; Cpr, cytochrome P450 reductase; Alas1, δ-aminolevulinic acid synthase; UDP-GA, UDP-glucuronic acid; PAPS, phosphoadenosine-5′-phosphosulfate; Papss2, PAPS synthase 2; Gclc, glutamate-cysteine ligase.
References
- Alnouti Y and Klaassen CD (2006) Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol Sci 93 242–255. [DOI] [PubMed] [Google Scholar]
- Ando H, Yanagihara H, Sugimoto K, Hayashi Y, Tsuruoka S, Takamura T, Kaneko S, and Fujimura A (2005) Daily rhythms of P-glycoprotein expression in mice. Chronobiol Int 22 655–665. [DOI] [PubMed] [Google Scholar]
- Baraldo M (2008) The influence of circadian rhythms on the kinetics of drugs in humans. Expert Opin Drug Metab Toxicol 4 175–192. [DOI] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, and Laudet V (2006) Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor α defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol 20 1715–1727. [DOI] [PubMed] [Google Scholar]
- Cheng X and Klaassen CD (2006) Regulation of mRNA expression of xenobiotic transporters by the pregnane X receptor in mouse liver, kidney, and intestine. Drug Metab Dispos 34 1863–1867. [DOI] [PubMed] [Google Scholar]
- Cheng X, Maher J, Chen C, and Klaassen CD (2005a) Tissue distribution and ontogeny of mouse organic anion transporting polypeptides (OATPs). Drug Metab Dispos 33 1062–1073. [DOI] [PubMed] [Google Scholar]
- Cheng X, Maher J, Dieter MZ, and Klaassen CD (2005b) Regulation of mouse organic anion-transporting polypeptides (OATPs) in liver by prototypical microsomal enzyme inducers that activate distinct transcription factor pathways. Drug Metab Dispos 33 1276–1282. [DOI] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, and Schibler U (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab 4 25–36. [DOI] [PubMed] [Google Scholar]
- Hartley DP and Klaassen CD (2000) Detection of chemical-induced differential expression of rat hepatic cytochrome P450 mRNA transcripts using branched DNA signal amplification technology. Drug Metab Dispos 28 608–616. [PubMed] [Google Scholar]
- Haus E and Smolensky M (2006) Biological clocks and shift work: circadian dysregulation and potential long-term effects. Cancer Causes Control 17 489–500. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Furihata T, Yaginuma Y, Yamamoto N, Koyano N, Fujii A, Nagahara Y, Satoh T, and Chiba K (2007) Genomic structure and transcriptional regulation of the rat, mouse, and human carboxylesterase genes. Drug Metab Rev 39 1–15. [DOI] [PubMed] [Google Scholar]
- Howell SR and Klaassen C (1991) Circadian variation of hepatic UDP-glucuronic acid and the glucuronidation of xenobiotics in mice. Toxicol Lett 57 73–79. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Otsuka S, Hiromasa T, Nakahama T, and Inouye Y (2004) Diurnal difference in CAR mRNA expression. Nucl Recept 2 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC and Lee SJ (1998) Temporal variation in hepatotoxicity and metabolism of acetaminophen in mice. Toxicology 128 53–61. [DOI] [PubMed] [Google Scholar]
- Klaassen CD and Lu H (2008) Xenobiotic transporters: ascribing function from gene knockout and mutation studies. Toxicol Sci 101 186–196. [DOI] [PubMed] [Google Scholar]
- Klaassen CD and Slitt AL (2005) Regulation of hepatic transporters by xenobiotic receptors. Curr Drug Metab 6 309–328. [DOI] [PubMed] [Google Scholar]
- Kotaka M, Onishi Y, Ohno T, Akaike T, and Ishida N (2008) Identification of negative transcriptional factor E4BP4-binding site in the mouse circadian-regulated gene Mdr2. Neurosci Res 60 307–313. [DOI] [PubMed] [Google Scholar]
- Levi F and Schibler U (2007) Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol 47 593–628. [DOI] [PubMed] [Google Scholar]
- Liu L and Klaassen CD (1996) Different mechanism of saturation of acetaminophen sulfate conjugation in mice and rats. Toxicol Appl Pharmacol 139 128–134. [DOI] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Callaghan TN, Cheng X, Cheung C, Gonzalez FJ, and Klaassen CD (2006) Alterations in transporter expression in liver, kidney, and duodenum after targeted disruption of the transcription factor HNF1α. Biochem Pharmacol 72 512–522. [DOI] [PubMed] [Google Scholar]
- Matsunaga N, Ikeda M, Takiguchi T, Koyanagi S, and Ohdo S (2008) The molecular mechanism regulating 24-hour rhythm of CYP2E1 expression in the mouse liver. Hepatology 48 240–251. [DOI] [PubMed] [Google Scholar]
- Mukai M, Lin TM, Peterson RE, Cooke PS, and Tischkau SA (2008) Behavioral rhythmicity of mice lacking AhR and attenuation of light-induced phase shift by 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Rhythms 23 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Tanaka Y, Nakano T, Adachi T, Tanaka H, Kaminuma T, and Ishikawa T (2006) Nuclear receptor-mediated transcriptional regulation in phase I, II, and III xenobiotic metabolizing systems. Drug Metab Pharmacokinet 21 437–457. [DOI] [PubMed] [Google Scholar]
- Noshiro M, Usui E, Kawamoto T, Kubo H, Fujimoto K, Furukawa M, Honma S, Makishima M, Honma K, and Kato Y (2007) Multiple mechanisms regulate circadian expression of the gene for cholesterol 7α-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J Biol Rhythms 22 299–311. [DOI] [PubMed] [Google Scholar]
- Oishi K, Shirai H, and Ishida N (2005) CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor α (PPARα) in mice. Biochem J 386 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson A and Ogilvie BW (2008) Biotransformation of xenobiotics, in Casarett and Doull's Toxicology: the Basic Science of Poisons (Klaassen CD ed) pp 161–304, McGraw-Hill, New York.
- Petrick JS and Klaassen CD (2007) Importance of hepatic induction of constitutive androstane receptor and other transcription factors that regulate xenobiotic metabolism and transport. Drug Metab Dispos 35 1806–1815. [DOI] [PubMed] [Google Scholar]
- Radzialowski FM and Bousquet WF (1968) Daily rhythmic variation in hepatic drug metabolism in the rat and mouse. J Pharmacol Exp Ther 163 229–238. [PubMed] [Google Scholar]
- Schnell RC, Bozigian HP, Davies MH, Merrick BA, Park KS, and McMillan DA (1984) Factors influencing circadian rhythms in acetaminophen lethality. Pharmacology 29 149–157. [DOI] [PubMed] [Google Scholar]
- Takiguchi T, Tomita M, Matsunaga N, Nakagawa H, Koyanagi S, and Ohdo S (2007) Molecular basis for rhythmic expression of CYP3A4 in serum-shocked HepG2 cells. Pharmacogenet Genomics 17 1047–1056. [DOI] [PubMed] [Google Scholar]
- Tukey RH and Strassburg CP (2000) Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol 40 581–616. [DOI] [PubMed] [Google Scholar]