Abstract
Accurate quantification of cytochrome P450 (P450) protein contents is essential for reliable assessment of drug safety, including the prediction of in vivo clearance from in vitro metabolism data, which may be hampered by the use of uncharacterized standards and existence of unknown allelic isozymes. Therefore, this study aimed to delineate the variability in absolute quantification of polymorphic CYP2D6 drug-metabolizing enzyme and compare immunoblot and nano liquid chromatography coupled to mass spectrometry (nano-LC/MS) methods in identification and relative quantification of CYP2D6.1 and CYP2D6.2 allelic isozymes. Holoprotein content of in-house purified CYP2D6 isozymes was determined according to carbon monoxide difference spectrum, and total protein was quantified with bicinchoninic acid protein assay. Holoprotein/total CYP2D6 protein ratio was markedly higher for purified CYP2D6.1 (71.0%) than that calculated for CYP2D6.1 Supersomes (35.5%), resulting in distinct linear calibration range (0.05–0.50 versus 0.025–0.25 pmol) that was determined by densitometric analysis of immunoblot bands. Likewise, purified CYP2D6.2 and CYP2D6.10 and the CYP2D6.10 Supersomes all showed different holoprotein/total CYP2D6 protein ratios and distinct immunoblot linear calibration ranges. In contrast to immunoblot, nano-LC/MS readily distinguished CYP2D6.2 (R296C and S486T) from CYP2D6.1 by isoform-specific proteolytic peptides that contain the altered amino acid residues. In addition, relative quantitation of the two allelic isozymes was successfully achieved with label-free protein quantification, consistent with the nominated ratio. Because immunoblot and nano-LC/MS analyses measure total P450 protein (holoprotein and apoprotein) in a sample, complete understanding of holoprotein and apoprotein contents in P450 standards is desired toward reliable quantification. Our data also suggest that nano-LC/MS not only facilitates P450 quantitation but also provides genotypic information.
Cytochrome P450 (P450) enzymes are the most important phase I drug-metabolizing enzymes responsible for the metabolic elimination of drugs in humans (Williams et al., 2004). Because of the striking species differences in P450-mediated drug metabolism (Lin, 1998; Gonzalez and Yu, 2006; McLaughlin et al., 2008), there remains high interest in quantitative prediction of in vivo hepatic drug clearance from in vitro metabolism data acquired from human liver microsomes, hepatocytes, and/or recombinant P450 enzymes according to physiology or population-based models (Obach, 1999; Houston and Galetin, 2003; Barter et al., 2007). Per se, a good understanding of the abundance of individual P450 proteins in human liver and small intestine and P450 pharmacogenetics (e.g., CYP2D6 and CYP2C9) is required for successful extrapolation of in vitro metabolic data to in vivo clearance parameters.
Immunoblot analysis is a conventional method in protein quantitation, and it has been widely used in assessing the abundance of individual P450s. Studies include the pioneering quantitative measurement of human hepatic (Guengerich and Turvy, 1991; Shimada et al., 1994) and intestinal (Paine et al., 2006) P450s, and the systemic characterization of ontogeny and possible sexual dimorphism of individual P450s in humans (Stevens et al., 2003, 2008; Wolbold et al., 2003) and CYP transgenes in mouse models (Yu et al., 2005; Cheung et al., 2006; Felmlee et al., 2008). However, immunoquantification determines total protein, holoprotein (active protein with prosthetic group heme properly incorporated) and apoprotein (inactive protein lacking of heme or nonholoprotein) for individual P450s in a given sample, whereas only holoprotein level is usually known for the standard and inevitably used for calibration. Indeed, the levels of holoprotein and apoprotein can be significantly different between the standards and samples, which causes the renowned discrepancy between CYP3A4 contents estimated with commercially available, unpurified recombinant enzymes as standards, and those assayed with purified enzymes (Perrett et al., 2007).
In terms of CYP2D6 quantitation, studies (Shimada et al., 1994; Paine et al., 2006) providing the basis for human hepatic and intestinal P450 “pie” all used unpurified CYP2D6 as standards, whose holoprotein/apoprotein levels were unfortunately unknown. Use of uncharacterized P450 standards including the purified proteins will certainly lead to inaccurate estimation of P450 contents in human tissues. Although the purified CYP2D6 enzymes from human liver microsomes have been shown to contain 6.3 to 38% of holoprotein (Gut et al., 1984; Distlerath et al., 1985), whether it represents the CYP2D6 holoprotein level in living human livers remains a question. First, substantial level of apoprotein may be removed or introduced during processing and purification. Second, polymorphic CYP2D6 consists of many different allelic isoforms that have significantly altered protein stability (Johansson et al., 1994), whereas the CYP2D6 genotypes of those liver donors were unknown. In addition, it is not clear whether one antibody developed for one isoform (usually the wild type) will react with another allelic isoform to the same degree in immunoblots.
Mass spectrometry (MS)-based methods not only reveal the identity of a protein analyte but also provide unparalleled sensitivity and selectivity for protein quantitation. Therefore, use of different MS techniques has become an indispensable approach for protein identification and quantification (Ong and Mann, 2005; Bantscheff et al., 2007). MS-based methods for absolute and relative quantification of P450 proteins are also emerging in recent years (Alterman et al., 2005; Jenkins et al., 2006; Duan et al., 2007; Lane et al., 2007). In a recent study, a nano-liquid chromatography (LC)/MS method has been adopted for proteomic analysis in our laboratory, which provides exceptional sensitivity and wealthy information of peptide sequence (Qu and Straubinger, 2005). This technique may be used to determine CYP2D6 abundance in a given sample and simultaneously identify allelic isoform-specific peptides, i.e., to provide genotype information.
Therefore, this study aimed to delineate the variability in immunoquantification of CYP2D6 caused by the use of different allelic isoforms from different sources, and to compare immunoblot and MS methods in relative quantification of CYP2D6.1 and CYP2D6.2 allelic isoforms. Our data showed that the immunoblot linear calibration ranges of purified and unpurified CYP2D6 allelic isozymes, which contain all levels of holoprotein and apoprotein, varied considerably. When P450 holoprotein content is only considered in the standard, level of the P450 in a sample will be markedly underestimated. In addition to the distinguishing of CYP2D6.1 from CYP2D6.2, which differ only in two amino acid residues, nano-LC/MS analysis successfully provided relative quantitation of the two allelic isoforms, and the result was consistent with the nominal value calculated from bicinchoninic acid (BCA) assay and immunoblot analysis.
Materials and Methods
Chemicals, Enzymes, and Other Reagents. CYP2D6.1 and CYP2D6.10 Supersomes and pooled human liver microsomes were purchased from BD Discovery Labware, Inc. (Woburn, MA). Iodoacetamide, formic acid, and l-1-tosylamido-2-phenylethyl chloromethyl ketone were bought from Sigma-Aldrich (St. Louis, MO). Trypsin was purchased from Promega (Madison, WI). In-house baculovirus-expressed CYP2D6.1, CYP2D6.2, and CYP2D6.10 were purified by Octyl-Sepharose, DEAE-Sepharose, and ceramic hydroxyapatite column chromatographies, as described previously (Yu et al., 2002). All the other chemicals used were of the highest analytical grade available.
Immunoblot Analysis. CYP2D6 samples containing 0.01 to 1.0 pmol of holoprotein were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) with a 12.5% resolving gel, and proteins were transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA). Membranes were first probed with monoclonal antibodies against human CYP2D6 (MAB-2D6; BD Discovery Labware, Inc.) and then incubated with peroxidase-conjugated goat anti-mouse IgG (Sigma-Aldrich), followed by enhanced chemiluminescence detection (Pierce Chemical, Rockford, IL) as described previously (Felmlee et al., 2008). Densitometric analysis was conducted using a Kodak Image Station (New Haven, CT). All the immunoblot analyses were conducted in duplicate or triplicate, and all the data were reproducible.
Determination of CYP2D6 Holoprotein and Total Protein Contents. Total protein concentrations were determined using the BCA Protein Assay Kit (Pierce Chemical) following the manufacturer's instructions. Holoprotein contents of CYP2D6 Supersomes were obtained from the manufacturer (BD Discovery Labware, Inc.). CYP2D6 holoprotein level of the purified CYP2D6 allelic isozymes was determined according to the carbon monoxide (CO) difference spectrum method (Omura and Sato, 1964), and total CYP2D6 concentration (holoprotein plus apoprotein; in molarity) was calculated according to its corresponding molecular mass after correction with the purity of CYP2D6 protein in each sample (90% for CYP2D6.1 and CYP2D6.2; 70% for CYP2D6.10) (Yu et al., 2002). Total protein levels were assayed in triplicate, and holoprotein contents were determined in duplicate, and mean values were obtained (Table 1).
TABLE 1.
Difference in CYP2D6 immunoblot linear calibration range and holoprotein/total CYP2D6 protein ratio between CYP2D6 Supersomes and in-house purified CYP2D6 allelic isoforms
Values represent mean of duplicate or triplicate measurements. Total protein concentration was determined using the BCA Protein Assay, and CYP2D6 holoprotein content was determined according to the CO difference spectrum. Total CYP2D6 concentration for in-house purified CYP2D6 isozyme was calculated according to its average molecular mass.
| Sample | Molecular Mass | Total Protein | CYP2D6 Holoprotein | Total CYP2D6 | Holoprotein/Total CYP2D6 Protein | Immunoblot Linear Range |
|---|---|---|---|---|---|---|
| Da | mg/ml | pmol/μl | pmol/μl | % | pmol | |
| CYP2D6.1 Supersomes | 55,769.5 | 3.20 | 1.00 | 2.82a | 35.5a | 0.025-0.25 |
| CYP2D6.10 Supersomes | 55,773.4 | 1.10 | 1.00 | 31.6a | 3.16a | 0.025-0.25 |
| CYP2D6.1 (in-house) | 55,769.5 | 0.60 | 6.90 | 9.72b | 71.0b | 0.05-0.50 (Fig. 1); 0.10-1.0 (Fig. 2) |
| CYP2D6.2 (in-house) | 55,730.4 | 0.60 | 5.00 | 9.72b | 51.4b | 0.10-1.0 |
| CYP2D6.10 (in-house) | 55,773.4 | 11.3 | 1.80 | 142b,c | 1.27b,c | 0.01-0.10 |
Estimated after compared with corresponding in-house CYP2D6 allelic isoforms (Fig. 1).
Values corrected with the purity of CYP2D6.
Low level of CYP2D6.10 holoprotein as a result of enzyme denaturation during purification (Yu et al., 2002).
Proteolytic Digestion. Ten microliters of CYP2D6.1 or CYP2D6.2 (total protein concentration 0.60 μg/μl for each sample) was diluted with 40 μlof Tris buffer (50 mM, pH = 8.6). Then Tris 2-carboxyethyl phosphine solution was spiked to a final concentration of 1 mM. The solution was heated at 95°C for 5 min to denature the protein and then cooled to room temperature. For alkylation of the cysteine residues, 1.8 μl of freshly prepared iodoacetamide solution (100 mM) was added. Then the mixture was incubated in darkness at room temperature for 30 min. l-1-Tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin was stored in 0.5% acetate acid and was activated by adding 3 volumes of 50 mM Tris buffer before the digestion. Activated trypsin was added to achieve a substrate/enzyme ratio of 20:1, and the reaction mixture was incubated at 37°C for 16 h. Proteolysis was stopped by the addition of 2 μl of formic acid. After centrifugation at 13,000 rpm for 5 min, the supernatants of triplicate digestions were subjected to nano-LC/MS analysis separately.
Nano-LC/MS and Protein Identification. A high-resolution nano-LC system, including two multidimensional LC nano pumps (GE Healthcare, Little Chalfont, Buckinghamshire, UK), a Spark Endurance autosampler (Emmen, The Netherlands), a laboratory-made valveless reverse-phase trap and nano-LC flow path, and two Vici10-port low-dead-volume valves, was used for protein separation. Programming of the nano-LC runs was performed on a Unicorn (GE Healthcare) coding platform. A laboratory-made dynamic-flow nanospray ion source was used to couple the nano-LC system to the LTQ XL linear ion trap tandem MS with an electron transfer dissociation (ETD) device attached (Thermo Fisher Scientific, San Jose, CA). In particular, the nano column was connected downstream of a metal T, on which the high voltage was applied (1.7–2.2 kV), and liquid junction approach was used to form the electrospray ionization Taylor cone on the uncoated spray tip. The MS was operated under data-dependent mode, in which one micro-scan cycle was composed of an MS1 survey scan, followed by one zoom MS1 scan with higher resolution, and then by six sequential dependent MS2 scans with collision-induced dissociation (CID) and ETD as activation method alternatively. The reaction time for ETD was set at 130 ms, and the chemical ionization source temperature was 230°C. Targeted value for negative ions was 6 × 105, and supplemental activation that uses a short CID of the charge-striped radical ion was used to further fragment the doubly charged precursors.
Tryptic samples were loaded on a reversed-phase peptide trap (5 mm × 300 μm i.d.) at a flow rate of 10 μl/min and washed with 3% of acetonitrile in 0.1% formic acid for 3 min to remove salts and other hydrophilic buffer components. Then the trap was switched online with a laboratory-packed reversed-phase nano C-18 column (3-μm particle size, reversed-phase particle packed in a 20-cm-long, 360 μm o.d., 75 μm i.d., and fused silica capillary ended with a noncoated 2-μm tampered tip). A shallow multistep gradient was used to resolve the samples, which included buffer A (3% acetonitrile in 0.1% formic acid) and buffer B (84% acetonitrile in 0.1% formic acid). One hundred percent buffer A was used for trap loading and the initial nano-LC equilibration. Then buffer B was increased linearly to 23% in 95 min, to 55% in the following 40 min, and to 73% in another 20 min, followed by 100% buffer B to flush the column for 15 min. On-column flow rates ranged from 180 to 260 nl/min. For CID, the raw data were converted data (DTA) files with zero-slack algorithm and Comblon filter algorithms to remove low-quality DTA files. The filtered DTA files were searched against the preindexed tryptic peptide database that was coded with known CYP2D6.1 sequences, with a number of possible post-translation modifications on specific amino acids. The resultant data were grouped into a search results file, and the peptide probability scores were calculated and a stringent probability filter (<0.01) was used to remove false-positive identification. To ensure the credibility of the result, a secondary group of filters was applied to the result: Xcorr > 2 for z = 1; Xcorr > 2.5 if z = 2; Xcorr > 3 if z = 3. Processing of ETD data was performed by Charger (Thermo Fisher Scientific) software and then searched for C and Z fragments using SEQUEST (Thermo Fisher Scientific). The filtering standard was the same as for CID as described above, except that a final score filter (>0.85) was used. For each of the identified peptides, a manual examination of the fragment pattern was performed to eliminate database searching artifacts.
Relative Quantification Based on Peptide Intensities by LC/MS. A label-free relative protein quantification approach was used to determine the relative level of the two CYP2D6 allelic isoforms. A commercial protein quantification software package, the DeCyder MS (GE Healthcare), was used to compare in triplicate the extracting ion currents (XICs) of the multiply charged peptide ions from CYP2D6.1 and CYP2D6.2. To facilitate the analysis, the retention times for XICs of the six runs were aligned before quantification. The background noise was subtracted by a uniform noise assumption, and any signal with signal/noise >20 was investigated for possible peptide candidates. For an MS1-matched peptide, its three-dimensional signal intensity (versus m/z and retention time) was determined. That is, the volume under the surface was obtained for the matched peptide by considering all the charge states after it was identified by both m/z calculation and XIC match. Of particular note, only frames (putative peptide signals) identified in all six samples (triplicate digestions of CYP2D6.1 and CYP2D6.2) were used for integration. After quantification, the DTA files derived from the MS2-dependent scans to the quantified precursors were exported and searched against protein database, as described above, for confirmatory identification of quantified peptides.
Results
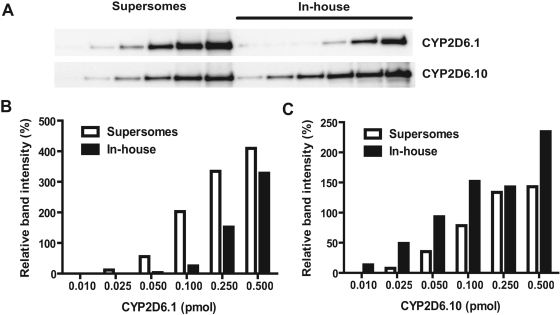
Immunoblot Analysis Revealed a Marked Difference in Linear Calibration Range for Individual CYP2D6 Allelic Isoforms from Different Sources. To investigate the source variability on the absolute quantitation of CYP2D6 protein, immunoblots of purified CYP2D6.1 versus CYP2D6.1 Supersomes and CYP2D6.10 versus CYP2D6.10 Supersomes were compared side-by-side, respectively (Fig. 1). Difference in band density is obvious between purified CYP2D6.1 and the same amount (based on CO difference spectrum) of unpurified CYP2D6.1 Supersomes. Quantitative densitometric analysis revealed that the two standards also showed distinct linear calibration ranges, 0.05 to 0.5 pmol for purified CYP2D6.1 versus 0.025 to 0.25 pmol for CYP2D6.1 Supersomes (Table 1). Likewise, the linear calibration ranges are markedly different between purified CYP2D6.10 (0.01–0.1 pmol) and CYP2D6.10 Supersomes (0.025–0.25 pmol). The results also indicate that different allelic isoforms (e.g., CYP2D6.1 or CYP2D6.10) from the same source (e.g., commercially available) could have distinct calibration curves. Depending on the standard selected, an overestimation or underestimation of CYP2D6 protein content is expected.
Fig. 1.
Immunoblot analysis (A) of unpurified (Supersomes) and purified (in-house) CYP2D6.1 and CYP2D6.10 reveals distinct quantitative densitometric responses for CYP2D6.1 (B) standards from different sources, and for CYP2D6.10 (C); 0.01, 0.025, 0.05, 0.1, 0.25, or 0.5 pmol holoprotein of individual allelic isozymes were loaded into each well, and proteins were separated by SDS-PAGE. Immunoblot analyses were conducted using a selective antibody against CYP2D6.
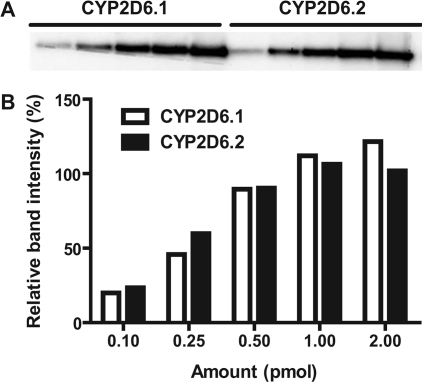
To further examine possible variability in CYP2D6 quantitation affected by CYP2D6 allelic isoforms, we used high-purity (>90% according to electrophoresis and LC), high-activity (≥5 pmol/μl according to CO difference spectrum or >8.0 pmol/mg protein) CYP2D6.1 and CYP2D6.2 for comparative immunoblot analysis (Fig. 2). It revealed that the same amount of CYP2D6.1 and CYP2D6.2 did exhibit similar band intensity, resulting in identical linear calibration ranges (Table 1). This indicates that immunoblot responses to the same monoclonal antibody do not differ between the two allelic isoforms and suggests that it is valid to quantify CYP2D6.2 and CYP2D6.1 with the same antibody. However, it does not exclude the possibility that other CYP2D6 allelic isoforms may respond differently from the wild-type CYP2D6.1.
Fig. 2.
Immunoblot (A) and densitometric analysis (B) of highly purified CYP2D6.1 and CYP2D6.2 suggests that CYP2D6 total protein levels are similar in the two samples; 0.1, 0.25, 0.5, 1.0, or 2.0 pmol holoprotein of each isozyme was loaded into each well, and proteins were separated by SDS-PAGE. Immunoblot analyses were carried out using a selective antibody against CYP2D6.
Considerable Variability in Holoprotein and Apoprotein Levels of Individual CYP2D6 Allelic Isoforms from Different Sources. To examine the causes of difference in immunoblot response of different CYP2D6 standards, we thoroughly determined CYP2D6 holoprotein and total protein contents for the purified CYP2D6 samples using CO difference spectrum method and BCA protein assay, respectively. According to the CYP2D6 total protein level, we further calculated the percentage of holoprotein for each sample (Table 1). It revealed that the holoprotein represents 71.0 and 51.4% of total (holoprotein plus apoprotein) CYP2D6 in purified CYP2D6.1 and CYP2D6.2 samples, respectively. Based on the comparative quantitative immunoblots (Fig. 1), we further estimated the total CYP2D6 level and the holoprotein/total CYP2D6 ratios for the CYP2D6.1 and CYP2D6.10 Supersomes. As expected, CYP2D6.1 Supersomes have much lower holoprotein/total CYP2D6 protein level (35.5%). In contrast, CYP2D6.10 Supersomes have relatively higher holoprotein/total CYP2D6 protein ratio than in-house CYP2D6.10 (3.16 versus 1.27%), which is presumably because of the denaturation of CYP2D6.10 during purification (Yu et al., 2002). Indeed, CYP2D6.10 has been known for its protein instability (Johansson et al., 1994) because of the change of N-terminal amino acid residue (P34S) critical for microsomal binding. Nevertheless, the large variability in holoprotein/apoprotein levels may explain the marked difference in immunoblot response of different allelic isoforms from different sources (Table 1).
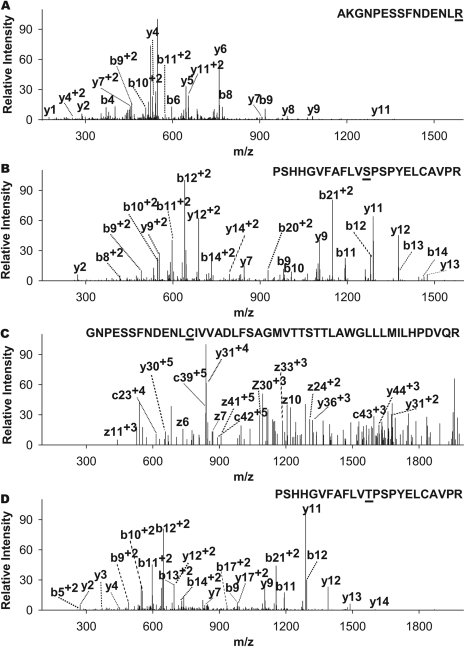
Identification and Relative Quantification of CYP2D6 Allelic Isoforms by Nano-LC/MS. To verify immunoblot quantitation, we used the nano-LC/MS for relative quantification of the two highly purified CYP2D6 allelic isoforms, CYP2D6.1 and CYP2D6.2. First, we identified isoform-specific tryptic peptides that distinguish CYP2D6.1 and CYP2D6.2 (R296C and S486T) from each other (Fig. 3). Because the majority of tryptic peptides from CYP2D6 allelic isoforms are rather polar, an optimized, shallow nano-LC gradient using low-organic mobile phase compositions was used to obtain relatively evenly distributed peptide peaks within the initial 90-min elution window (data not shown) for comprehensive peptide identification. Two complementary techniques, CID and ETD, were used alternatively for data-dependent fragmentation of the tryptic peptides to achieve higher sequence coverage. Consequently, 92% of the predicated amino acid residues were identified for CYP2D6.1 with high confidence and 89% for CYP2D6.2. Two expected tryptic peptides from CYP2D6.2 (Fig. 3), which contain the altered amino acid residues (Cys296 and Thr486), were readily identified by either CID or ETD. It is notable that the Cys296 residue in CYP2D6.2 precluded tryptic cleavage at C-terminal of this position, in contrast to the Arg296 in CYP2D6.1. As a result, a longer tryptic peptide containing 46 amino acid residues was produced. Because of the larger number of residues, this peptide was highly charged and thus not suitable for CID analysis as a result of a possible mobile proton impediment mechanism. Indeed, this peptide was not identified by CID but detected by ETD (Fig. 3C) that is especially efficient for long and highly charged peptide ions. In contrast, the other three isoform-specific tryptic peptides were identified not only by CID (Fig. 3, A, B, and D) but also by ETD (data not shown).
Fig. 3.
LC/tandem MS identification of the difference in amino acid residues between CYP2D6.2 (R296C and S486T) and CYP2D6.1 allelic isozymes. A, CID MS identification of AKGNPESSFNDENLR from CYP2D6.1. B, CID MS identification of PSHHGVFAFLVSPSPYELCAVPR from CYP2D6.1. C, ETD MS identification of GNPESSFNDENL-CIVVADLFSAGMVTTSTTLAWGLLLMILH-PDVQR from CYP2D6.2. D, CID MS identification of PSHHGVFAFLVTPSPYELCAVPR from CYP2D6.2.
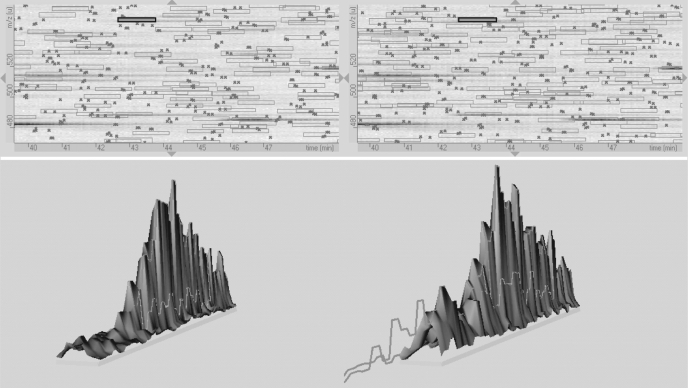
Label-free protein quantification, which was performed with the DeCyder MS software package, was then conducted to compare the relative level of CYP2D6 in CYP2D6.1 and CYP2D6.2 samples. To ensure the accuracy of the quantification, the MS1 ion channel for quantification was obtained by a stand-alone high-resolution LTQ scan (full width at half maximum = 0.1 Th), which greatly improved the success of ion match by DeCyder MS. Second, a three-dimensional integration was used to calculate the volume under the surface of individual signals (intensity versus the retention time and m/z) that include all the isotopic forms and at all the charge states from the putative peptide precursor ions of CYP2D6.1 and CYP2D6.2. A representative relative quantification of the two isozymes using a matched peptide (FGDIVPLGVTHMTSR) is shown in Fig. 4. The data indicate that the relative amount of total CYP2D6 protein between the purified CYP2D6.1 and CYP2D6.2 is around 1:1.2 (Table 2), in agreement with the nominal ratio (1:1) derived from BCA protein assay and selective CYP2D6 immunoblots (Fig. 2).
Fig. 4.
Illustration of relative quantification of CYP2D6.2 versus CYP2D6.1 by DeCyder MS using a matched tryptic peptide (FGDIVPLGVTHMTSR; in the highlighted areas) existing in both isozymes. Top, zoom view of paired MS1 signals (m/z versus retention time); bottom, the three-dimensional integration of the representative peptide at all the charge states.
TABLE 2.
Relative level of total CYP2D6 between the purified CYP2D6.2 and CYP2D6.1 calculated from representative matched peptides by DeCyder MS
After characterization with BCA protein assay, CO difference spectrum, and immunoblot analysis, equal amount (holoprotein + apoprotein) of CYP2D6.2 and CYP2D6.1 was used for the analysis.
| Peptide IDa | Retention Time | Peptide Sequence | CYP2D6.2/CYP2D6.1 Ratio | Xcorr |
|---|---|---|---|---|
| min | ||||
| 130 | 34.2 | R.MTWDPAQPPR.D | 1.22 | 3.14 |
| 556 | 43.2 | R.FGDIVPLGVTHMTSR.D | 1.11 | 3.38 |
| 473 | 43.3 | K.DEAVWEKPFR.F | 1.23 | 3.31 |
| 253 | 50.2 | R.LLDLAQEGLK.E | 1.34 | 2.61 |
| 214 | 52.1 | R.VQQEIDDVIGQVR.R | 1.39 | 5.25 |
| 205 | 60.0 | R.FGDIVPLGVTHMTSR.D | 1.22 | 2.69 |
| 476 | 63.1 | K.AVSNVIASLTCGR.R | 1.45 | 2.79 |
| 112 | 87.2 | K.GTTLITNLSSVLK.D | 1.28 | 4.20 |
| 141 | 93.3 | R.PPVPITQILGFGPR.S | 1.19 | 2.61 |
| Mean ± S.D. | 1.27 ± 0.11 |
A unique ID for each matched peptide assigned by the DeCyder MS “Pepmatch” module.
Discussion
Although human hepatic and intestinal P450 contents are readily quantified by immunoblot analysis with form-selective antibodies, discrepancy is obvious between the use of purified and unpurified P450 standards, which may hamper accurate quantitative prediction of in vivo drug clearance from in vitro metabolic data. Therefore, the current study compared the difference in immunoblot response and linear calibration range between in-house purified CYP2D6 allelic isoforms and commercially available CYP2D6 Supersomes, which are basically governed by the total CYP2D6 content, including both holoprotein and apoprotein. Nano-LC/MS analysis not only provided accurate quantitation of CYP2D6.1 and CYP2D6.2 allelic isoforms but also proved the identity of each protein by delineation of form-specific tryptic peptides.
Immunoquantification of a P450 determines both holoprotein and apoprotein of the P450, i.e., total P450 protein. In contrast, not the total P450 protein but holoprotein level is usually known for the P450 standards, especially those commercially available membrane preparations. Disparity has been noted that CYP3A4 contents assayed with purified CYP3A4 standards are significantly higher than the levels determined with unpurified standards (Perrett et al., 2007). It is notable that P450 quantitation is more complex than that assumed, even if the holoprotein/apoprotein ratios were the same in a standard and a sample. The holoprotein level of each purified P450 from human livers summarized by Perrett et al. (2007) truly indicates the holoprotein level of corresponding P450 in the purified sample or represents protein stability and difficulty in processing, rather than the level of P450 (e.g., CYP2D6) holoprotein in living human tissues. Less or unstable P450 isozymes may be partially denatured or inactivated as processed. On the other hand, because of distinct lipophilicity or binding affinity, some denatured P450 protein (apoprotein) may be separated from holoprotein on certain columns during purification. Therefore, whether all or some of the translated P450 enzymes in living human tissues are holoproteins awaits further investigation.
When compared with purified CYP2D6.1, the same amount of active CYP2D6.1 Supersomes that are often used for quantification of CYP2D6 (Paine et al., 2006; Stevens et al., 2008) shows stronger response to the same monoclonal antibody (Fig. 1) and thus different linear calibration range in immunoblot quantitation (Table 1). This indicates that the abundance of CYP2D6 will be markedly underestimated when CYP2D6.1 Supersomes are used as the standard and its apoprotein level is not considered. Agreeing with findings of Perrett et al. (2007), the reason is that the CYP2D6.1 Supersomes consist of much lower level of holoprotein (holoprotein/total CYP2D6 protein, 35.5%) than the in-house purified CYP2D6.1 (71.0%). In contrast, we have found that the holoprotein levels are also largely variable (Table 3) in purified recombinant CYP2D6 enzymes that were reported in the literature (Gillam et al., 1995; Kempf et al., 1995; Imaoka et al., 1996; Modi et al., 1996; Yu et al., 2002; Ng et al., 2003; Rowland et al., 2006). Most of them do show relatively higher holoprotein levels (CYP2D6 holoprotein/total protein, >50%), whereas the others have comparable ratios (14–40%) as CYP2D6.1 Supersomes (36%) or purified CYP2D6 from human tissues (6.3–38%) (Gut et al., 1984; Distlerath et al., 1985). Of particular note, those P450s purified from human donors would be superb quantitation standards when they are characterized properly.
TABLE 3.
CYP2D6 holoprotein levels (percentage of total protein) in the purified recombinant CYP2D6 enzymes reported in the literature
Average molecular mass of each protein was calculated according to its corresponding amino acid sequence described in that article unless otherwise indicated.
| Protein | Molecular Mass | Theoretical CYP2D6 Content | Experimental CYP2D6 Content | Holoprotein Level | Expression System | Reference |
|---|---|---|---|---|---|---|
| Da | mg/ml | mg/ml | % | |||
| CYP2D6m-Δ 21 | 53,262.2 | 18.8 | 10.0 | 53.2 | Escherichia coli | Gillam et al., 1995 |
| [His]6-CYP2D6-Δ 25 | 55,535.7 | 18.0 | 7.12a, 2.67b | 39.6a, 14.8b | E. coli | Kempt et al., 1995 |
| CYP2D6-Δ | 48,500c | 20.6 | 12.2 | 59.2 | Yeast | Imaoka et al., 1996 |
| CYP2D6-Ext | 56,235.1 | 17.8d, 20.0c | 18.3 | 91.5d | Baculovirus (insect cell) | Modi et al., 1996 |
| CYP2D6.1 | 55,769.5 | 17.9 | 11.5 | 64.2 | Baculovirus (insect cell) | Yu et al., 2002 |
| CYP2D6m | 55,647.3 | 18.0 | 12.5 | 69.4 | E. coli | Ng et al., 2003 |
| CYP2D6m-Δ-[His]4 | 53,705.7 | 18.6 | 11.0 | 59.1 | E. coli | Roland et al., 2006 |
Purified without detergent.
Purified with detergent.
Values reported in corresponding papers.
Value we calculated according to its molecular mass.
With the advances of MS in relative and absolute protein quantitation, some MS methods have also been developed for quantitative analysis of P450 isozymes (Alterman et al., 2005; Jenkins et al., 2006; Duan et al., 2007; Lane et al., 2007). Our nano-LC/MS method has proved to provide excellent sensitivity in protein quantification (Qu and Straubinger, 2005). In the current study, the ratio of total CYP2D6 between CYP2D6.1 and CYP2D6.2 samples determined by label-free quantification of form-matched peptides (Table 2; Fig. 4) separated by nano-LC is close to the nominated value calculated from total protein assay (Table 1) and selective CYP2D6 immunoblots (Fig. 2). Although MS methods usually provide excellent sensitivity (e.g., attomole to femtomole), they share the same limitation as immunoblot analyses in determining the total (active and inactive) level of the analyzed protein. Therefore, use of uncharacterized P450 standards, whose holoprotein levels are determined only, would not provide reliable quantitation of the P450 in human tissues.
In addition to relative quantification of P450 proteins, Lane et al. (2007) have nicely shown the application of nano-LC/MS to peptide identification for individual P450s. Although the majority of P450s of interest were unambiguously identified, that method was unable to distinguish two highly homologous isozymes, CYP2A4 and CYP2A5, from each other. To identify CYP2D6.1 and CYP2D6.2 allelic isozymes that differ only in two amino acid residues, our nano-LC/MS method uses two complementary fragmentation techniques, CID and ETD, and an optimal peptide separation condition. In turn, higher sequence coverage (92% for CYP2D6.1 and 89% for CYP2D6.2) has been achieved. Among them, the CYP2D6.2 form-specific tryptic peptide derived as a result of the C296R change consists of 46 amino acids and may not be suitable for CID, whereas it has been readily identified by ETD (Fig. 3). Our finding suggests that CYP2D6 genotyping, which is usually accomplished by the analysis of human DNA or mRNA samples (Chou et al., 2003; Meijerman et al., 2007), may be also achieved by direct analysis of CYP2D6 protein in human biopsy using appropriate MS methods.
In summary, MS-based methods not only provide accurate quantification of P450 proteins but also identify allelic isoform-specific peptides. Because immunoblot and MS analyses determine both inactive and active protein for a P450 enzyme, it is essential to use fully characterized P450 standards toward better understanding of P450 contents in humans and reliable prediction of in vivo clearance from in vitro metabolic data.
This work was supported in part by the National Institutes of Health (NIH)/National Institute on Drug Abuse [Grant R01DA021172].
The LTQ/ETD MS system was obtained from NIH/National Center for Research Resources.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.024166.
ABBREVIATIONS: P450, cytochrome P450; MS, mass spectrometry; LC, liquid chromatography; BCA, bicinchoninic acid; PAGE, polyacrylamide gel electrophoresis; CO, carbon monoxide; ETD, electron transfer dissociation; CID, collision-induced dissociation; XIC, extracting ion current; DTA, data.
References
- Alterman MA, Kornilayev B, Duzhak T, and Yakovlev D (2005) Quantitative analysis of cytochrome p450 isozymes by means of unique isozyme-specific tryptic peptides: a proteomic approach. Drug Metab Dispos 33 1399–1407. [DOI] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, and Kuster B (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389 1017–1031. [DOI] [PubMed] [Google Scholar]
- Barter ZE, Bayliss MK, Beaune PH, Boobis AR, Carlile DJ, Edwards RJ, Houston JB, Lake BG, Lipscomb JC, Pelkonen OR, et al. (2007) Scaling factors for the extrapolation of in vivo metabolic drug clearance from in vitro data: reaching a consensus on values of human microsomal protein and hepatocellularity per gram of liver. Curr Drug Metab 8 33–45. [DOI] [PubMed] [Google Scholar]
- Cheung C, Yu AM, Chen CS, Krausz KW, Byrd LG, Feigenbaum L, Edwards RJ, Waxman DJ, and Gonzalez FJ (2006) Growth hormone determines sexual dimorphism of hepatic cytochrome P450 3A4 expression in transgenic mice. J Pharmacol Exp Ther 316 1328–1334. [DOI] [PubMed] [Google Scholar]
- Chou WH, Yan FX, Robbins-Weilert DK, Ryder TB, Liu WW, Perbost C, Fairchild M, de Leon J, Koch WH, and Wedlund PJ (2003) Comparison of two CYP2D6 genotyping methods and assessment of genotype-phenotype relationships. Clin Chem 49 542–551. [DOI] [PubMed] [Google Scholar]
- Distlerath LM, Reilly PE, Martin MV, Davis GG, Wilkinson GR, and Guengerich FP (1985) Purification and characterization of the human liver cytochromes P-450 involved in debrisoquine 4-hydroxylation and phenacetin O-deethylation, two prototypes for genetic polymorphism in oxidative drug metabolism. J Biol Chem 260 9057–9067. [PubMed] [Google Scholar]
- Duan X, Chen X, Yang Y, and Zhong D (2007) Precolumn derivatization of cysteine residues for quantitative analysis of five major cytochrome P450 isoenzymes by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 21 3234–3244. [DOI] [PubMed] [Google Scholar]
- Felmlee MA, Lon HK, Gonzalez FJ, and Yu AM (2008) Cytochrome P450 expression and regulation in CYP3A4/CYP2D6 double transgenic humanized mice. Drug Metab Dispos 36 435–441. [DOI] [PubMed] [Google Scholar]
- Gillam EM, Guo Z, Martin MV, Jenkins CM, and Guengerich FP (1995) Expression of cytochrome P450 2D6 in Escherichia coli, purification, and spectral and catalytic characterization. Arch Biochem Biophys 319 540–550. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ and Yu AM (2006) Cytochrome P450 and xenobiotic receptor humanized mice. Annu Rev Pharmacol Toxicol 46 41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP and Turvy CG (1991) Comparison of levels of several human microsomal cytochrome P-450 enzymes and epoxide hydrolase in normal and disease states using immunochemical analysis of surgical liver samples. J Pharmacol Exp Ther 256 1189–1194. [PubMed] [Google Scholar]
- Gut J, Gasser R, Dayer P, Kronbach T, Catin T, and Meyer UA (1984) Debrisoquine-type polymorphism of drug oxidation: purification from human liver of a cytochrome P450 isozyme with high activity for bufuralol hydroxylation. FEBS Lett 173 287–290. [DOI] [PubMed] [Google Scholar]
- Houston JB and Galetin A (2003) Progress towards prediction of human pharmacokinetic parameters from in vitro technologies. Drug Metab Rev 35 393–415. [DOI] [PubMed] [Google Scholar]
- Imaoka S, Yamada T, Hiroi T, Hayashi K, Sakaki T, Yabusaki Y, and Funae Y (1996) Multiple forms of human P450 expressed in Saccharomyces cerevisiae. Systematic characterization and comparison with those of the rat. Biochem Pharmacol 51 1041–1050. [DOI] [PubMed] [Google Scholar]
- Jenkins RE, Kitteringham NR, Hunter CL, Webb S, Hunt TJ, Elsby R, Watson RB, Williams D, Pennington SR, and Park BK (2006) Relative and absolute quantitative expression profiling of cytochromes P450 using isotope-coded affinity tags. Proteomics 6 1934–1947. [DOI] [PubMed] [Google Scholar]
- Johansson I, Oscarson M, Yue QY, Bertilsson L, Sjöqvist F, and Ingelman-Sundberg M (1994) Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol 46 452–459. [PubMed] [Google Scholar]
- Kempf AC, Zanger UM, and Meyer UA (1995) Truncated human P450 2D6: expression in Escherichia coli, Ni(2+)-chelate affinity purification, and characterization of solubility and aggregation. Arch Biochem Biophys 321 277–288. [DOI] [PubMed] [Google Scholar]
- Lane CS, Wang Y, Betts R, Griffiths WJ, and Patterson LH (2007) Comparative cytochrome P450 proteomics in the livers of immunodeficient mice using 18O stable isotope labeling. Mol Cell Proteomics 6 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH (1998) Applications and limitations of interspecies scaling and in vitro extrapolation in pharmacokinetics. Drug Metab Dispos 26 1202–1212. [PubMed] [Google Scholar]
- McLaughlin LA, Dickmann LJ, Wolf CR, and Henderson CJ (2008) Functional expression and comparative characterization of nine murine cytochromes P450 by fluorescent inhibition screening. Drug Metab Dispos 36 1322–1331. [DOI] [PubMed] [Google Scholar]
- Meijerman I, Sanderson LM, Smits PH, Beijnen JH, and Schellens JH (2007) Pharmacogenetic screening of the gene deletion and duplications of CYP2D6. Drug Metab Rev 39 45–60. [DOI] [PubMed] [Google Scholar]
- Modi S, Paine MJ, Sutcliffe MJ, Lian LY, Primrose WU, Wolf CR, and Roberts GC (1996) A model for human cytochrome P450 2D6 based on homology modeling and NMR studies of substrate binding. Biochemistry 35 4540–4550. [DOI] [PubMed] [Google Scholar]
- Ng PS, Imaoka S, Hiroi T, Osada M, Niwa T, Kamataki T, and Funae Y (2003) Production of inhibitory polyclonal antibodies against cytochrome P450s. Drug Metab Pharmacokinet 18 163–172. [DOI] [PubMed] [Google Scholar]
- Obach RS (1999) Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: an examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos 27 1350–1359. [PubMed] [Google Scholar]
- Omura T and Sato R (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239 2370–2378. [PubMed] [Google Scholar]
- Ong SE and Mann M (2005) Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol 1 252–262. [DOI] [PubMed] [Google Scholar]
- Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, and Zeldin DC (2006) The human intestinal cytochrome P450 “pie.” Drug Metab Dispos 34 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett HF, Barter ZE, Jones BC, Yamazaki H, Tucker GT, and Rostami-Hodjegan A (2007) Disparity in holoprotein/apoprotein ratios of different standards used for immunoquantification of hepatic cytochrome P450 enzymes. Drug Metab Dispos 35 1733–1736. [DOI] [PubMed] [Google Scholar]
- Qu J and Straubinger RM (2005) Improved sensitivity for quantification of proteins using triply charged cleavable isotope-coded affinity tag peptides. Rapid Commun Mass Spectrom 19 2857–2864. [DOI] [PubMed] [Google Scholar]
- Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, et al. (2006) Crystal structure of human cytochrome P450 2D6. J Biol Chem 281 7614–7622. [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, and Guengerich FP (1994) Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270 414–423. [PubMed] [Google Scholar]
- Stevens JC, Hines RN, Gu C, Koukouritaki SB, Manro JR, Tandler PJ, and Zaya MJ (2003) Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther 307 573–582. [DOI] [PubMed] [Google Scholar]
- Stevens JC, Marsh SA, Zaya MJ, Regina KJ, Divakaran K, Le M, and Hines RN (2008) Developmental changes in human liver CYP2D6 expression. Drug Metab Dispos 36 1587–1593. [DOI] [PubMed] [Google Scholar]
- Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, and Ball SE (2004) Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos 32 1201–1208. [DOI] [PubMed] [Google Scholar]
- Wolbold R, Klein K, Burk O, Nüssler AK, Neuhaus P, Eichelbaum M, Schwab M, and Zanger UM (2003) Sex is a major determinant of CYP3A4 expression in human liver. Hepatology 38 978–988. [DOI] [PubMed] [Google Scholar]
- Yu A, Kneller BM, Rettie AE, and Haining RL (2002) Expression, purification, biochemical characterization, and comparative function of human cytochrome P450 2D6.1, 2D6.2, 2D6.10, and 2D6.17 allelic isoforms. J Pharmacol Exp Ther 303 1291–1300. [DOI] [PubMed] [Google Scholar]
- Yu AM, Fukamachi K, Krausz KW, Cheung C, and Gonzalez FJ (2005) Potential role for human cytochrome P450 CYP3A4 in estradiol homeostasis. Endocrinology 146 2911–2919. [DOI] [PubMed] [Google Scholar]