Abstract
Previous studies have shown that Msx proteins control gene transcription predominantly through repression mechanisms. However, gene expression studies using either the gain-of-function or the loss-of-function mutants revealed many gene targets whose expression require functional Msx proteins. To date, investigations into the mechanisms of Msx-dependent trans-activation have been hindered by the lack of a responsive promoter. Here, we demonstrated the usefulness of the mouse Hspa1b promoter in probing Msx-dependent mechanisms of gene activation. We showed that Msx protein activates Hspa1b promoter via its C-terminal domain. The activation absolutely depends on the HSEs and physical interactions between Msx proteins and Heat shock factors may play a contributing role.
Keywords: Msx1, Msx2, transcriptional activator, Hsp70, Hspa1b, promoter analysis, protein-protein interaction, homeodomain, Heat shock factor, HSF1, HSF2, Heat Shock Elements, HSE, cotransfection, mutagenesis
Introduction
Msx homeoproteins are major players in orchestrating morphogenetic events that contribute to craniofacial, brain and eye development [1–10]. They are known to affect transcription by forming repressive protein-protein heteromeric complexes [11–20]. However, growing experimental evidence also point to their positive role in activating transcription. In a recent study, Msx1 was shown to physically interact with Pax9 and enhance the ability of Pax9 to transactivate Msx1 and Bmp4 promoters when Msx1 was co-transfected at a low concentration [21]. Msx1-deficiency leads to significantly reduced expression of Bmp4 (40), Fgf3, Lef1, Ptc, Dlx2, and syndecan-1 in the dental mesenchyme [22–23]. Overexpression of Msx2 in the optic vesicle was shown to induce Bmp7 expression [9]. Msx2 mutants showed decreased expression of Runx2, Alkaline phosphatase (Alp), bone sialoprotein (Bsp), osteocalcin (Osc), and Pthr in long bones [6]. In the floor plate of embryonic neural tube Shh-promoter directed expression of Msx1 in transgenic mice induced Ngn2 expression [24]. In cultured myoblasts, Msx1 was capable of inducing CyclinD1 expression [25]. In the Msx1/Msx2 double mutant, expression of cell cycle inhibitor p27, Fgf10 and Hand1/2 were suppressed in the developing heart [10]. Together, these results suggest that Msx proteins can activate transcription.
In this report, we present first evidence demonstrating that Msx1 and Msx2 are potent transcriptional activators in the context of the Hspa1b (formerly Hsp70) promoter. We showed that the activator activity is mediated by the C-terminal domain of Msx proteins and that physical interactions between Msx and Heat shock factors may contribute to Hspa1b promoter activities.
Material and Methods
Plasmid construction
The Hspa1b-LacZ reporter plasmid (pHspPTlacZpA) was kindly provided by Dr. Janet Rossant (University of Toronto, Canada) [26]. To construct serial deletions and introduce point mutations into the Hspa1b promoter, DNA fragments were generated by performing PCR and were subsequently inserted into the XhoI/NcoI sites of pHspPTlacZpA. The reporter plasmid pHsp-90 was constructed by removing DNA sequences between StuI and SmaI sites of pHspPTlacZpA.
To generate Msx1 and Msx2 mutant expression plasmids, PCR fragments were ligated in-frame to the 3x Flag tag in the pIRES-hrGFP-1a vector (Stratagene, La Jolla, CA). To create the Dlx5-homeodomain Msx C-terminal domain chimeric construct, a PCR fragment spanning the Dlx5 homeodomain was inserted in-frame upstream of pMsx1-236–303. Msx2-GFP and Msx2-233-GFP were constructed by inserting PCR fragments into XhoI and BamHI sites of pEGFP-c1 (Clontech, Pola Alto, CA). All DNA modifications were verified by sequencing.
Cell culture and DNA transfection
C2C12 myoblast cells and αTN4 mouse lens epithelial cells were cultured in DMEM supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). Before transfection, cells were seeded in 12 well plates. When cells reached a density of approximately 2×105 cells per well, transfections were performed using Lipofectamine and Plus reagent according to manufacturer’s protocol (Invitrogen, Carlsbad, CA). Each well received a total of 800ng plasmid DNA that consisted of 100ng of individual Hspa1b-lacZ reporter plasmid, 300ng Msx expression plasmid, 50ng CMV-luciferase used as the internal standard to control for transfection efficiency and a variant amount of pIRES-hrGFP-1a to adjust for the deficit in total amount of DNA. Each transfection was performed in triplicates.
Dual β-galactosidase and luciferase assays
Luciferase and beta-galatosidase assays were performed using the Dual-light Assay kit (Applied Biosystems, Bedford, MA) following manufacturers recommendations with a few modifications. Twenty-four hours after transfection, cell lysis was performed directly in 12-well plates by adding 80μl lysis buffer per well. Cell lysates were transferred into microfuge tubes and cell debris pelleted by centrifugation. 10μl of individual lysates were used in the subsequent enzymatic activity assay. Reporter activities as a function of photon emission were measured using a Berthold luminometer (Lumat LB9507, Berthold).
Immunoprecipitation and immunoblotting assays
C2C12 cells were transfected with pMsx1-3xflag, pMsx1- Δ;AD-3xflag, or pMsx2-3xflag when cells reached a density of about 60% confluency. Twenty-four hours post-transfection, cells were harvested and placed in 1ml of cold Lysis Buffer (50mM Tris-HCl pH 8.0, 150mM NaCl;1% NP-40) containing 1X Protease Inhibitor Cocktail (Sigma-Aldrich, St. Louis, MO) and incubated on ice for 1 hour. Cell debris was spun down at 10,000×g for 15 minutes at 4°C. To preclear the cell lysate, either 50μl anti-rabbit IgG beads (eBioscience, San Diego, CA) or anti-rat IgG beads (Sigma-Aldrich, St. Louis, MO) was added into the cell lysate and was incubated on ice for 30 minutes. After centrifugation, 4μg rabbit anti-HSF1 (Stressgen, Victoria, BC, Canada) or rat anti-HSF2 (Lab Vision, Fremont, CA) antibody was added into the supernatant and was subjected to overnight incubation at 4°C. On the next day, an additional 50μl of anti-rabbit IgG beads or anti-rat IgG beads was added into the sample and incubated for 60 minutes. Beads were then spun down by centrifugation at 10,000×g for 1 minute. Supernatant was completely removed and beads were washed 4 times each with 1000μl of TBS Buffer (50mM Tris-HCl pH 8.0, 150mM NaCl, 0.1% tween 20). After the last wash, 50μl of 1X SDS Reducing Sample Buffer (6% SDS, 50mM DTT, 25mM Tris base PH 6.5, 10% glycrol, bromphenol blue) was added to resuspend the bead pellet and then boiled for 10 minutes. Boiled beads were spun down by centrifugation and the supernatant was loaded onto a PAGE gel and analyzed by Western blotting. Flag-tagged Msx proteins were detected using a mouse monoclonal anti-Flag M2 antibody conjugated to HRP (1:2000 dilution; Sigma-Aldrich, St. Louis, MO).
To immunoprecipitate Flag-tagged Msx fusion protein, 40 μl anti-Flag (M2) antibody conjugated agarose-beads (Sigma-Aldrich, St. Louis, MO) were added into precleared lysates and allowed to incubate overnight at 4°C. Beads were pelleted by centrifugation, washed and resuspended in 50 μl 1X SDS Reducing Sample Buffer. Immunoprecipitated proteins were then released from antibody conjugated beads by boiling and analyzed by Western blotting using a rabbit anti-HSF1 antibody (1:5000 dilution; Stressgen, Victoria, BC, Canada) or a rat anti-HSF2 antibody (1:1000 dilution; Lab Vision, Fremont, CA).
Results
Msx enhances transcriptional activity of the Hspa1b promoter
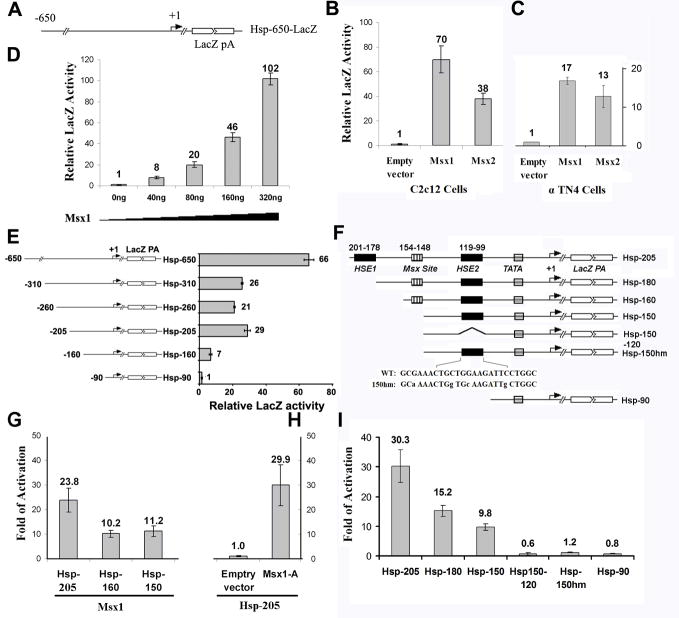
Hspa1b promoter has been widely used as a basal promoter for studying enhancer activity in transgenic mice (47). We found that a 650bp DNA promoter fragment of the mouse Hspa1b gene (Hsp-650) responded positively to Msx1 and Msx2 induction in C2C12 myoblast cells, α-TN4 lens epithelial cells (Figure 1A, 1B, 1C) as well as in primary mice MEFs (data not shown). Co-transfection of the 650bp Hsp70-lacZ reporter plasmid (Hsp-650) with increasing amounts of Msx1 expression plasmid revealed that Msx1 can directly influence Hspa1b promoter activity in a gene dosage dependent manner (Figure 1D).
FIG. 1. Activation of Hspa1b promoter by the Msx1 and Msx2.
(A) Schematic of the 650bp Hspa1b promoter-lacZ reporter construct used in transient transfection assays. (B) The Hspa1b promoter responded robustly to the induction by Msx1 and Msx2 in C2C12 cells. (C) Cotransfection of Msx1 with the Hspa1b-reporter construct also resulted in the transcriptional induction of the Hspa1b promoter in αTN4 mouse epithelial cell line. (D) The Hspa1b promoter showed dose-dependent response to Msx1. (E) A series of promoter deletion constructs (left) was made to map cis regulatory sequences that responded to Msx induction (right). A 205bp Hspa1b promoter fragment was found to be sufficient in responding to the induction by the Msx1. C2C12 cells were co-transfected with 100 ng of individual Hspa1b deletion-reporter plasmids and 300 ng Msx1-3xFlag expression plasmids or an empty control plasmid. Beta-galactosidase activities were normalized to an internal control expressing luciferase. Each transfection was performed in triplicates with the standard error shown. (F). A schematic representations of Hspa1b promoter reporter deletion constructions. HSE1 and HSE2 corresponding to Heat shock factor binding sites and a Msx binding site are shown. Mutations introduced into the HSE2 site are indicated by lower-case letters. The striped rectangle denotes the position of the TATA box. (G) The Msx binding site is not required for the induction of Hspa1b promoter activity by Msx1. Removal of the Msx consensus binding site didn’t affect the transcriptional response of the Hspa1b promoter as transcriptional activities between the Hsp-160 and Hsp-150 failed to show significant difference. (H) Cotransfection of a non-DNA binding Msx1 mutant (Msx1-A) did not alter the ability of the mutant protein to transactivate the 205bp Hspa1b promoter-reporter. All transfections were performed in triplicates. (I). Deletion of the distal HSE (Hsp-180) lead to a 50% reduction in Msx1-stimulated transcriptional activity; removal of both HSEs (Hsp150-120) or mutagenized HSE2 (Hsp-150hm) abolished the Msx1-dependent promoter activity. All transfections were performed in triplicates.
To determine cis-regulatory sequences that are required for Msx-dependent trans-activation, a series of overlapping deletions were examined. Two major cis-regulatory domains were identified: the first one is embedded between nucleotides -650 and -310 and the other spans between nucleotides -205 and -90. Removal of 445bp from the 5′ end of Hsp-650 (Hsp-205) resulted in an about two fold reduction in the Msx1-dependent transcriptional activity (Figure 1A, 1E). Further deletion of 50bp from nucleotide -205 (Hsp-160) resulted in an additional two to three fold reduction in transcriptional activity, hinting the existence of additional positive cis-regulatory sequences between nucleotides -205 and -156 (Figure 1E). Further deletion of 66bp from -156 (Hsp-90) nearly abolished Msx-dependent transcriptional activity (Figure 1E).
Msx1-dependent trans-activation does not require direct DNA binding by Msx1
To further define cis-acting elements between nucleotides -205 and -90 that may function in synergy with Msx1, we focused on two heat shock elements (HSE, -201 to -178, -119 to -99), one potential Msx consensus binding site (-154 to -148) (Figure 1F). To determine if DNA binding is required for transcriptional activation by Msx1, the Msx consensus binding site was removed from Hsp-160 to generate the Hsp-150 reporter construct. Co-transfection of either the Hsp-160 or the Hsp-150 produced similar transcriptional response (Figure 1G). To further rule out the possibility of cryptic binding activity, a non-DNA binding Msx1 mutant (Msx1-A) [19] was co-transfected with Hsp-205. The Msx1-A mutant did not diminish Hsp-205 promoter activity (Figure 1H). Together these results indicated that Msx1 can function as a transcriptional activator independent of its DNA binding activity.
Msx1-dependent trans-activation requires heat shock response elements
A deletion of the distal HSE (Hsp-180) resulted in a 50% reduction in Msx1-dependent transcriptional activity (compare Hsp-205 with Hsp-180, Figure 2I). Removal of both Heat Shock elements (Hsp150-120) totally abolished Msx1-dependent activation (Figure 2I). To identify the specific role of HSE in Msx1 transactivation function, point mutations were induced into the HSE of Hsp-150 (Figure 1F). As shown in Figure 1I, the mutant HSE (Hsp-150hm) completely abolished the Msx1-dependent promoter activation. These results showed the critical importance of the HSE in mediating Msx1-dependent transcriptional response.
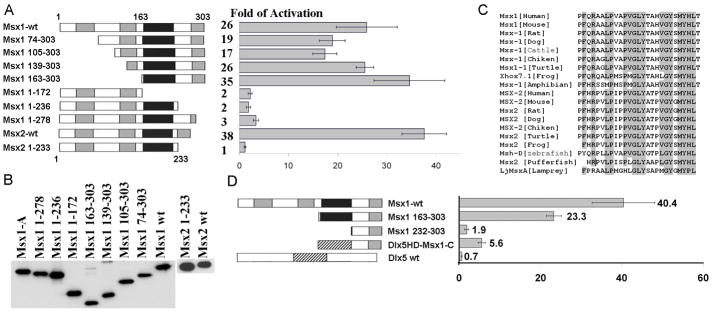
FIG. 2. The transcriptional activation activity resides in the C-terminal domain of Msx proteins.
(A) Schematic representations of the full-length and deletion mutants of Msx1 and Msx2. Areas that shaded gray represent Msx conserved regions and areas that shaded black correspond to the homeodomain. Removal of the C-terminal domain in both Msx1 and Msx2 protein leaded to a significant reduction in reporter activity. (B) Msx mutants that were transfected into C2C12 cells were stably translated. Flag-tagged Msx1 and Msx2 wild type and mutants proteins were detected on western blots using the anti-Flag antibody. (C) The C-terminal domains are highly conserved among Msx1 and Msx2 proteins in vertebrates. Identical amino acids were shaded in gray. (D) Schematic of replacement of Msx homeodomain (black) with Dlx5 homeodomain (striped rectangle). Removal of the homeodomain (Msx1 232-303) resulted in a significant reduction in the magnitude of transcriptional stimulation in comparison to Msx1 163-303 which contains the homeodomain. Replacement of the Msx1 homeodomain with the Dlx5 homeodomain restored the activation potency. 100 ng of Hsp205-LacZ reporters with 300 ng Msx1 mutant derivatives, Msx1-Dlx5HD or pIRES-hrGFP-1a were co-transfected into C2C12 cells. Transfections were performed in triplicates, and the standard error is shown.
The Msx1 activation domain resides in its C-terminus
Since the transcriptional activation function of Msx1 and its DNA binding function are separable, it is most likely that these functional domains are mirrored on its primary structure. To identify a functional domain that is required for transcriptional trans-activation, a series of Msx1 deletion mutants was constructed (Figure 2A, 2B). These deletion mutants were co-transfected into C2C12 cells along with the Hsp-205 reporter. Removal of N-terminal domains (Msx1 163-303) did not alter Hsp-205 reporter activities (Figure 2A); whereas, deletion of the homeodomain (Msx1 1-172) or removal of 67 to 25 residues (Msx1 1-236 or Msx1 1-278) beyond the homeodomain caused a major reduction in transcriptional activity (Figure 2A), indicating the critical importance of the C-terminus in conferring transactivation activity.
The transactivation domain is highly conserved among Msx1 and Msx2 proteins
ClustalW multi-sequence alignment showed high conservation of a 26 amino acid sequence in the C-termini among vertebrate Msx1 and Msx2 proteins (Figure 2C). To demonstrate their conserved role in activating transcription, 34 amino acids of the C-terminus were removed from the mouse Msx2 protein (Msx2 1-233). This resulted in a 38-fold reduction in transcriptional activity in comparison to the full length Msx2 (Figure 2A, 2B).
Homeodomain acts in synergy with the C-terminus in transactivation
Previous studies have shown that the Msx homeodomain tethers to a variety of protein complexes [14–19]. To determine the role of the homeodomain in stimulating transcription, we replaced the Msx1 homeodomain with the Dlx5 homeodomain (DLX5HD-Msx1-C) since Dlx5 can form a heterodimer with Msx proteins through their respective homeodomains [14]. Co-transfection of DLX5HD-Msx1-C with the Hsp-205 reporter resulted in three fold more activation in comparison to the homeodomain deletion mutant Msx1 163-303; whereas Dlx5 alone failed to induce promoter activity (Figure 2D). Together, these results demonstrate that the activation is mediated by the Msx C-terminal domain and that the homeodomain contributes to the magnitude of activation.
Msx proteins stimulate Hspa1b promoter activity by interacting with Heat Shock Factors
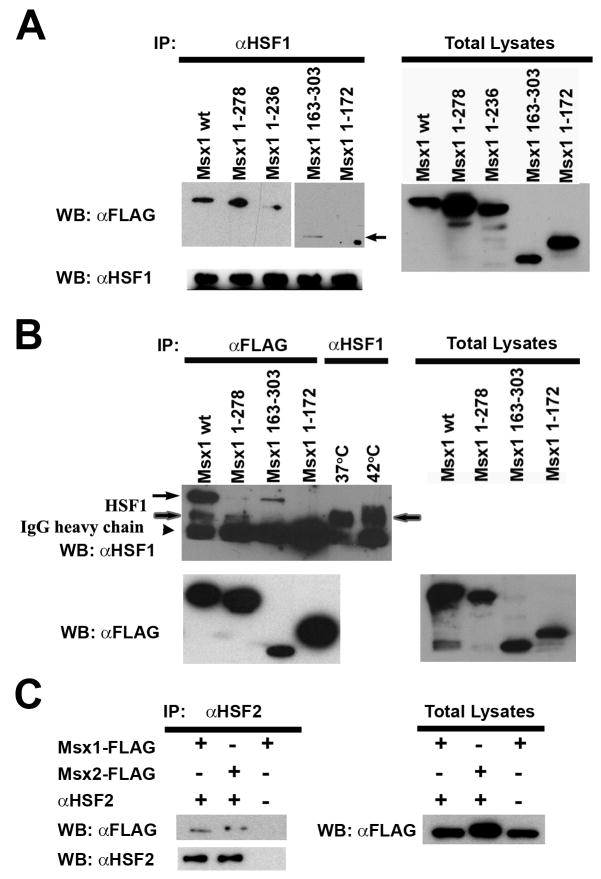
Because Msx1-dependent activation of Hspa1b promoter requires heat shock elements, it is most likely that Msx1 activates transcription by forming activating complexes with Heat Shock Factors, such as HSF1 and/or HSF2. Co-immunoprecipitation was performed to demonstrate physical interactions between Msx proteins and HSF proteins in vivo. The full-length Msx1 (Msx1 wt) and the C-terminal deletion or N-terminal deletion which contain the homeodomain (Msx1 1-278, Msx1 1-236, and Msx1 163-303) bind HSF1 while HSF1 failed to pull down the homeodomain deletion mutant (Msx1 1-172) (Figure 3A). Immuno-precipitation using the anti-Flag antibody and probed western blots utilizing the anti-Hsf1 antibody showed that the Msx2 wt, Msx1 N-terminal deletion mutant (Msx1 163-303) also pulled down a unique 95kDa HSF1 band; whereas the Flag-tagged C-terminal deletion mutant (Msx1 1-278) pulled down only the native 75kDa HSF1 (Figure 3B). These results demonstrated that the physical contact between Msx1 and Hsf1 is mediated by the Msx homeodomain.
FIG. 3. Msx proteins and HSFs physically interact in vivo.
(A) Msx1 binds to HSF1 through its homeodomain. Msx1 mutants that contain the homeodomain without either the N-terminal domain (Msx1 163-303) (arrow) or the C-terminal domain (Msx1-1-236) immunoprecipitated endogenous HSF1, while a mutant without the homeodomain (Msx1 1-172) failed to immunoprecipitate endogenous HSF1. (B) The Msx1 wildtype protein (Msx1 wt) and the Msx1 163-303 mutant which contains both the homeodomain and the C-teriminal domain immunoprecipitated a 95kD protein (arrow) that cross reacts to the anti-HSF1 antibody and the native 75kD HSF1 (shaded arrow). However, the Msx1 1-278 and Msx1 1-172 that lack the C-terminal domain failed to immunoprecipitate the 95kD protein species. The banding patterns of HSF1 under heated (42°C) and unheated conditions (37°C) provide a molecular weight reference for the endogenous HSF1. (C) Flag-tagged Msx1 and Msx2 can be immunoprecipitated using the anti-HSF2 antibody.
Besides binding to HSF1, we found that both Msx1 and Msx2 also physically interact with HSF2. To demonstrate this, immunoprecipitation was performed using a rat anti-HSF2 antibody. Immunoprecipitated proteins were blotted and probed with the anti-Flag antibody. HSF2 pulled down both Msx1 and Msx2 (Figure 3C).
Discussion
This study provides new insight into transcriptional regulatory functions of Msx1 and Msx2 proteins. We demonstrated for the first time that Msx proteins functions as transcriptional transactivators in the context of the murine Hspa1b promoter and that Msx-dependent transactivation does not require its DNA binding function although the homeodomain is required for maximizing transactivation activity. Msx2 shares similar activities in activating the Hspa1b promoter, indicating that these proteins are functionally equivalent in the context of the Hspa1b promoter activation.
Domain deletion analysis placed the activation domain in the C-termini of Msx proteins. The C-terminal domain consists of 26 amino acid residues and is uniquely conserved among Msx1 and Msx2 members of the broader msh gene family. Interestingly, a recent study identified binding of Msx1 to Pias1 through this domain [29]. Pias1 recruits Msx1 to nuclear periphery and enable it to bind to the distal enhancer on the MyoD gene to suppress its transcription. Elimination of the Pias1 interaction domain, the equivalence of the C-terminal domain in the present study, also abolishes Msx1’s suppressor activity and rids its ability to inhibit myoblast differentiation, demonstrating its critical importance in controlling cell differentiation [29].
Our analysis of the Hspa1b promoter also uncovered a critical need for heat shock factor binding sites and implicated Heat Shock Factors in Msx-dependent activation of the Hspa1b promoter. Co-immunoprecipitation demonstrated physical binding between Msx1/Msx2 and HSF1/HSF2. These physical interactions are mediated by the Msx homodomain. However, siRNA blocking of HSF1 translation resulted in less than two fold reduction in Hspab1b promoter activity (data not shown), suggesting involvement of additional co-activators that recognize HSEs.
The fact that Msx1 and Msx2 can stimulate transcription of Hspa1b promoter suggests that Msx proteins are capable of activating transcription of gene targets. Further characterization of Msx co-activator complexes in the context of Hspa1b promoter shall shed new light on additional partners and transcriptional regulatory mechanisms that involve Msx proteins in directing developmental processes.
Acknowledgments
We thank Dr. Henry Fong and Dr. Micheal Paine for advice and suggestions. This work was supported by the National Institutes of Health grants DE12941 and EY015417.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jabs EW, Muller U, Li X, Ma L, Luo W, Haworth IS, Klisak I, Sparkes R, Warman ML, Mulliken JB, et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 2.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 3.Houzelstein D, Cohen A, Buckingham ME, Robert B. Insertional mutation of the mouse Msx1 homeobox gene by an nlacZ reporter gene. Mech Dev. 1997;65:123–133. doi: 10.1016/s0925-4773(97)00065-8. [DOI] [PubMed] [Google Scholar]
- 4.Liu YH, Tang Z, Kundu RK, Wu L, Luo W, Zhu D, Sangiorgi F, Snead ML, Maxson RE. Msx2 gene dosage influences the number of proliferative osteogenic cells in growth centers of the developing murine skull: a possible mechanism for MSX2-mediated craniosynostosis in humans. Dev Biol. 1999;205:260–274. doi: 10.1006/dbio.1998.9114. [DOI] [PubMed] [Google Scholar]
- 5.Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 6.Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, Peters H, Tang Z, Maxson R, Maas R. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 7.van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhao X, Hu Y, St Amand T, Zhang M, Ramamurthy R, Qiu M, Chen Y. Msx1 is required for the induction of Patched by Sonic hedgehog in the mammalian tooth germ. Dev Dyn. 1999;215:45–53. doi: 10.1002/(SICI)1097-0177(199905)215:1<45::AID-DVDY5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 9.Wu LY, Li M, Hinton DR, Guo L, Jiang S, Wang JT, Zeng A, Xie JB, Snead M, Shuler C, Maxson RE, Jr, Liu YH. Microphthalmia resulting from MSX2-induced apoptosis in the optic vesicle. Invest Ophthalmol Vis Sci. 2003;44:2404–2412. doi: 10.1167/iovs.02-0317. [DOI] [PubMed] [Google Scholar]
- 10.Chen YH, Ishii M, Sun J, Sucov HM, Maxson RE., Jr Msx1 and Msx2 regulate survival of secondary heart field precursors and post-migratory proliferation of cardiac neural crest in the outflow tract. Dev Biol. 2007;308:421–437. doi: 10.1016/j.ydbio.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 11.Towler DA, Rutledge SJ, Rodan GA. Msx-2/Hox 8.1: a transcriptional regulator of the rat osteocalcin promoter. Mol Endocrinol. 1994;8:1484–1493. doi: 10.1210/mend.8.11.7877617. [DOI] [PubMed] [Google Scholar]
- 12.Catron KM, Zhang H, Marshall SC, Inostroza JA, Wilson JM, Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol Cell Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catron KM, Wang H, Hu G, Shen MM, Abate-Shen C. Comparison of MSX-1 and MSX-2 suggests a molecular basis for functional redundancy. Mech Dev. 1996;55:185–199. doi: 10.1016/0925-4773(96)00503-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen MM, Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bendall AJ, Ding J, Hu G, Shen MM, Abate-Shen C. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development. 1999;126:4965–4976. doi: 10.1242/dev.126.22.4965. [DOI] [PubMed] [Google Scholar]
- 16.Zhou YL, Lei Y, Snead ML. Functional antagonism between Msx2 and CCAAT/enhancer-binding protein alpha in regulating the mouse amelogenin gene expression is mediated by protein-protein interaction. J Biol Chem. 2000;275:29066–29075. doi: 10.1074/jbc.M002031200. [DOI] [PubMed] [Google Scholar]
- 17.Newberry EP, Latifi T, Battaile JT, Towler DA. Structure-function analysis of Msx2-mediated transcriptional suppression. Biochemistry. 1997;36:10451–10462. doi: 10.1021/bi971008x. [DOI] [PubMed] [Google Scholar]
- 18.Ichida F, Nishimura R, Hata K, Matsubara T, Ikeda F, Hisada K, Yatani H, Cao X, Komori T, Yamaguchi A, Yoneda T. Reciprocal roles of MSX2 in regulation of osteoblast and adipocyte differentiation. J Biol Chem. 2004;279:34015–34022. doi: 10.1074/jbc.M403621200. [DOI] [PubMed] [Google Scholar]
- 19.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa T, Kapadia H, Feng JQ, Raghow R, Peters H, D’Souza RN. Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J Biol Chem. 2006;281:18363–18369. doi: 10.1074/jbc.M601543200. [DOI] [PubMed] [Google Scholar]
- 21.Woloshin P, Song K, Degnin C, Killary AM, Goldhamer DJ, Sassoon D, Thayer MJ. MSX1 inhibits myoD expression in fibroblast x 10T1/2 cell hybrids. Cell. 1995;82:611–620. doi: 10.1016/0092-8674(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann HM, Catron KM, van Wijnen AJ, McCabe LR, Lian JB, Stein GS, Stein JL. Transcriptional control of the tissue-specific, developmentally regulated osteocalcin gene requires a binding motif for the Msx family of homeodomain proteins. Proc Natl Acad Sci U S A. 1994;91:12887–12891. doi: 10.1073/pnas.91.26.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- 24.Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development. 1998;125:4325–4333. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- 25.Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 26.Hu G, Lee H, Price SM, Shen MM, Abate-Shen C. Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development. 2001;128:2373–2384. doi: 10.1242/dev.128.12.2373. [DOI] [PubMed] [Google Scholar]
- 27.Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Mivechi NF. HSF-1 interacts with Ral-binding protein 1 in a stress-responsive, multiprotein complex with HSP90 in vivo. J Biol Chem. 2003;278:17299–17306. doi: 10.1074/jbc.M300788200. [DOI] [PubMed] [Google Scholar]
- 29.Lee H, Quinn JC, Prasanth KV, Swiss VA, Economides KD, Camacho MM, Spector DL, Abate-Shen C. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 2006;20:784–794. doi: 10.1101/gad.1392006. [DOI] [PMC free article] [PubMed] [Google Scholar]