Abstract
Quinones represent an important class of endogenous compounds such as neurotransmitters and coenzyme Q10, electrophilic xenobiotics, and environmental toxicants that have known reactivity based on their ability to redox cycle and generate oxidative stress, as well as to alkylate target proteins. It is likely that topological, chemical, and physical features combine to determine which proteins become targets for chemical adduction. Chemical-induced post-translational modification of certain critical proteins causes a change in structure/function that contributes to the toxicological response to chemical exposure. In this study, we have identified a number of proteins that are modified by quinone-thioethers after administration of 2-(glutathion-S-yl)HQ. Parallel one-dimensional gel electrophoresis was performed, and the Coomassie-stained gel was aligned with the corresponding Western blot, which was probed for adductions. Immunopositive bands were then subjected to trypsin digestion and analyzed via liquid chromatography/tandem mass spectrometry. The proteins that were subsequently identified contained a higher than average (9.7 versus 5.5%) lysine content and numerous stretches of lysine run-ons, which is a presumed electrophile binding motif. Approximately 50% of these proteins have also been identified as targets for electrophilic adduction by a diverse group of chemicals by other investigators, implying overlapping electrophile adductomes. By identifying a motif targeted by electrophiles it becomes possible to make predictions of proteins that may be targeted for adduction and possible sites on these proteins that are adducted. An understanding of proteins targeted for adduction is essential to unraveling the toxicity produced by these electrophiles.
Proteins have long been recognized as being critical targets of chemicals that produce acute tissue toxicities and cancer. Moreover, the adverse effects of many drugs are frequently mediated via their metabolic activation to reactive electrophilic metabolites, which subsequently bind covalently to proteins. Such reactive metabolites usually have low electron density and react with molecular centers of high electron density (i.e., nucleophiles). Target proteins for adduction usually contain strong nucleophilic sites, including cysteine thiols, lysine amines, histidine imidazoles, and protein N-terminal amines, which are readily attacked by reactive species (Guengerich et al., 2001; Casini et al., 2002). Other proteins contain weaker nucleophilic sites, including methionine sulfur, arginine guanidinium, tyrosine phenols, serine and threonine hydroxyls, and aspartate and glutamate carboxyls.
Attempts to identify protein targets of reactive electrophilic metabolites have been complicated because 1) proteins have tremendously diverse structures and properties, 2) unmodified proteins are frequently present in excess, and 3) radiolabeled chemicals of sufficiently high specific activity are often unavailable or prohibitively expensive. Antibody-based approaches to adduct detection have revealed a handful of targets for several chemicals, but only in a few cases has adduction been clearly defined at the level of amino acid sequence (Stewart et al., 2007; Ikehata et al., 2008). Thus, identifying specific sites of reactive chemical/metabolite-mediated modification within proteins has been extremely challenging. However, recent developments in mass spectrometry (MS) ionization methods and instrumentation now make possible the rapid, high-throughput, and sensitive analysis of proteins.
Nonetheless, identification of multiple and novel post-translational modifications remains a major challenge. In particular, chemical-induced post-translational modifications are usually of low abundance and thus not readily detectable by standard proteomics protocols. We reported the comparative analysis of proteolytic digests from control and treated proteins by matrix-assisted laser desorption ionization MS to target differences caused by chemical-induced modifications, without initial assumption as to type or residue localization (Person et al., 2003). Differences between modified and unmodified digest MS spectra highlighted peptides of interest, with targeted high-performance liquid chromatography (HPLC) coupled to electrospray ionization-tandem MS subsequently used to fragment peptides, and finally the amino acid sequence and type of modification were determined manually (Person et al., 2003). This in vitro model system consisted of cytochrome c adducted with 1,4-benzoquinone (BQ), an electrophilic metabolite of benzene. De novo sequencing identified a novel cyclized diquinone adduct as the major reaction product, targeting lysine and histidine residues at two specific locations on the protein surface (Person et al., 2003). A metabolite of BQ, 2-(glutathion-S-yl)-1,4-benzoquinone, a nephrotoxic quinol-thioether (QT) metabolite of BQ, also forms covalent adducts with cytochrome c, targeting several histidine and lysine residues (Person et al., 2005). The initial reaction product rearranges to a disubstituted cyclic quinone species via quinol amine linkages preferentially found at Lys25 to Lys27 and Lys86 to Lys87. These two sites were also identified as targets of BQ adduct formation (Person et al., 2003). Cyclic reaction products are preferentially formed at these sites presumably because of the presence of multiple basic residues present in a conformationally flexible region. Furthermore, when cytochrome c is reacted with 2-(N-acetylcystein-S-yl)-1,4-benzoquinone an additional six peptides were identified as containing lysine adducts residing on very basic portions of the protein, generally immediately adjacent to another basic residue (Fisher et al., 2007).
We subsequently proposed that reactive electrophiles may preferentially adduct specific motifs within proteins [electrophile binding motifs (EBMs)] that facilitate the chemical adduction reaction (Fisher et al., 2007). To examine this hypothesis, we used an established Eker rat (Tsc-2EK/+) model of chemical-induced nephrotoxicity and nephrocarcinogenicity (Lau et al., 2001), combined with the development of proteomics methods (Kleiner et al., 1998a,b; Person et al., 2003), to isolate adducted proteins and to ascertain features that predispose proteins to chemical adduction. We report that 2-(glutathion-S-yl)-1,4-benzoquinone-adducted proteins contain either lysine residues flanking a potentially nucleophilic amino acid (KXK) or two lysine residues preceded, or followed, by a nucleophilic amino acid (XKK or KKX). Knowledge of such preferential EBMs is a prerequisite for determining the potential biological/toxicological consequences of chemical adduction to proteins.
Materials and Methods
Chemicals. Actin, ammonium bicarbonate, dithiothreitol, formic acid, iodoacetamide, and trifluoroacetic acid were purchased from Sigma-Aldrich (St. Louis, MO), and 2-(glutathion-S-yl)-hydroquinone (MGHQ) was synthesized in our laboratory as described previously (Lau et al., 1988). Antibody sources were as follows: rabbit anti-2-Br-6-(N-acetylcystein-S-yl)hydroquinone (anti-2-BrHQ-NAC) in house (Kleiner et al., 1998b); peroxidase-labeled goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA); and antiactin and peroxidase-labeled goat anti-mouse IgG (Calbiochem, San Diego, CA). Enhanced chemiluminescent reagent (ECL) and Hyperfilm ECL were purchased from GE Healthcare (Little Chalfont, Buckinghamshire, UK). Sequencing grade trypsin was purchased from Promega (Madison, WI).
Treatment of Animals. Male Eker rats (Tsc-2EK/+) were obtained from a breeding colony at Science Park Research Division of the University of Texas M.D. Anderson Cancer Center (Houston, TX) and were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited facility with access to food and water ad libitum. Rats were administered MGHQ [400 μmol/kg, tail vein i.v. (0.1 ml/100 g)] or vehicle [0.1 ml/100 g, phosphate-buffered saline (PBS), pH 7.4] and then individually housed in metabolism cages where urine was collected on ice. Animals were euthanized by CO2 asphyxiation, and kidneys were removed at 2-h postinjection. Urine was extracted from the bladder and pooled with the excreted urine, and γ-glutamyltranspeptidase (γ-GT) activity was assayed as a marker of nephrotoxicity (Lau et al., 2001). One unit of γ-GT activity is defined as 1 μmol p-nitroanilide formed/min at 37°C. Significant increases in urinary γ-GT activity in treated rats were observed when compared with control rats (32.3 ± 7.1 versus 1.5 ± 0.8 units of total γ-GT over 2 h). The outer stripe of the outer medulla (OSOM), which is a target of MGHQ toxicity, was subsequently excised from the kidney and stored until further analysis. Tissues from animals receiving the same experimental treatments were pooled. The cytosolic, microsomal, mitochondrial, nuclear, and plasma membrane fractions of the OSOM were isolated by differential centrifugation, according to established methodology (Kleiner et al., 1998b).
Western Analysis of MGHQ-Adducted Proteins. Subcellular fractions were subjected to duplicate one-dimensional SDS-polyacrylamide gel electrophoresis (PAGE). Proteins (100–150 μg/lane) were resolved on 10 to 13% acrylamide gels at 100 V; one gel was stained with Coomassie Blue, and its duplicate was electrophoretically transferred to nitrocellulose. The nitrocellulose Western blot was incubated overnight at 4°C with rabbit anti-2-BrHQ-NAC and then incubated with peroxidase-labeled goat anti-rabbit IgG for 1.5 h at room temperature, followed by ECL solution for 1 min (Kleiner et al., 1998b). The bands in the Coomassie Blue-stained gel, corresponding to the immunoreactive bands in the Western blot, were excised and subjected to in-gel tryptic digestion.
In-Gel Tryptic Digestion of Immunopositive Proteins. In-gel tryptic digests were conducted according to Rosenfeld et al. (1992) with slight modifications. In brief, gels were dried with acetonitrile and reduced with dithiothreitol, followed by alkylation with iodoacetamide to prevent free thiol groups from cross-linking. Gel bands were dried and rehydrated in ammonium bicarbonate, then trypsinized at 37°C overnight. The resulting peptides were extracted from the gels with 5% formic acid/acetonitrile (50:50, v/v).
Liquid Chromatography/Tandem Mass Spectrometry Analysis of In-Gel Tryptic Digests. Peptides were separated via microbore HPLC (MAGIC 2002; Michrom BioResources, Auburn, CA) on a 0.5 × 50-mm MAGIC MS C18 column (5-μm, 200-Å pore size) using a mobile phase of acetonitrile/water/acetic acid/trifluoroacetic acid, A (2:98:0.1:0.02, v/v) and B (10:90:0.09: 0.02, v/v), with a gradient of 5 to 65% B over 30 min at a flow rate of 20 μl/min. The HPLC was coupled on-line with an electrospray-ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) set to a positive mode spray voltage of 3.5 kV and capillary temperature of 200°C. Maximum injection time was 50 ms for full scan and 200 ms for five tandem mass spectrometry (MS/MS) microscans. Dependent data setting was performed with a default charge of 2, an isolation width of 2 atomic mass units (amu), activation amplitude of 35%, activation time of 30 ms, and a minimal signal of 50,000 ion counts. Global dependent data settings were as follows: reject mass width of 1 amu, dynamic exclusion enabled, exclusion mass width of 3 amu, repeat count of 3, repeat duration of 1 min, and exclusion duration of 1 min. Scan event series included one full scan with mass range 400 to 2000 Da, followed by one dependent MS/MS scan of the most intense ion.
Identification of Sequenced Immunopositive Tryptic Peptides. Peptides were sequenced using the SEQUEST algorithm, incorporated into Thermo Fisher Scientific BIOWORKS software, to search and correlate the MS/MS spectra with amino acid sequences in the OWL protein database from the National Center for Biotechnology Information [(NCBI) Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST/]. Peptide sequences from the urinary excreted proteins were also identified using the open-source search engine X!Tandem, which, similar to SEQUEST, searches and correlates the MS/MS spectra with amino acid sequences in a user-specified NCBI database (ipi.RAT.v3) (Craig and Beavis, 2003).
Identification of Potential Motif for Quinol-Thioether Adduction of Immunopositive Proteins. EBMs were identified using Motif_Hunter (trademark to SSL, University of Arizona, Tucson, AZ). FASTA sequences were obtained from the Swiss-Prot database input into Motif_Hunter and were searched for the motifs KK and KXK. The outputs from the program were visually verified for accuracy, and data were compiled. Lysine occurrence (%) was also calculated by Motif_Hunter.
Results
Recognition and Isolation of Adducted Proteins. An immunogen was synthesized by coupling 2-BrHQ-NAC to keyhole-limpet hemocyanin (KLH) (Kleiner et al., 1998b). The brominated, l-N-acetylcysteine analog was selected because it provided a more stable substrate for the initial linkage reaction. Anti-2-BrHQ-NAC-KLH antibodies were raised in rabbits and purified by affinity chromatography (Kleiner et al., 1998b). Antibody binding to the 2-BrHQ-NAC epitope was confirmed by competitive enzyme-linked immunosorbent assay with 2-BrHQ-NAC-bovine serum albumin (Kleiner et al., 1998b). Affinity purified anti-2-BrHQ-NAC-KLH antibodies specifically recognized proteins from the kidneys of Eker rats (Tsc-2EK/+) treated with MGHQ. Immunoreactive protein bands were more intense in cytosolic extracts obtained from the OSOM compared with cortical cystolic extracts (data not shown), which is consistent with the fact that the OSOM is the initial target of MGHQ toxicity. Previous immunohistochemical analysis of tissue obtained 2 h after administration of MGHQ revealed immunopositive staining localized to the S3 segment of the renal proximal tubules at the corticomedullary junction along the medullary rays and in OSOM, areas that correlate with the subsequent region of necrosis (Kleiner et al., 1998a). Consequently, the OSOM and cortex were excised, and the nuclei, mitochondria, plasma membrane, microsomes, and cytosol-enriched fractions were isolated by differential centrifugation.
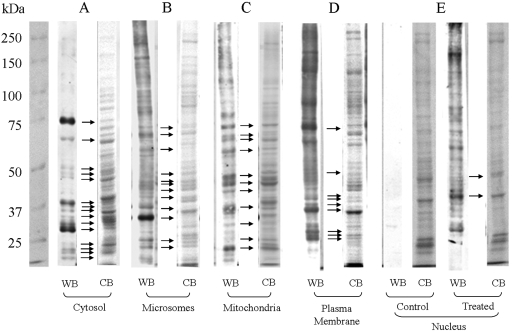
Western Blot Analysis Followed by In-Gel Digestion and Liquid Chromatography/MS/MS Protein Identification. The Coomassie Blue staining pattern of proteins from treated and control animals was identical for each of the five subcellular fractions. The Coomassie Blue staining of cytosol (Fig. 1A), microsomes (Fig. 1B), mitochondria (Fig. 1C), plasma membrane (Fig. 1D), and nuclei were distinct (Fig. 1E). The control tissue of all the fractions was immunonegative, and for illustrative purposes, only the nuclei from untreated controls are shown (Fig. 1E). The pattern and the intensity of the immunostained proteins in the MGHQ-treated rat kidney fractions were markedly different from the corresponding Coomassie Blue staining patterns. Immunoreactive proteins were excised (Fig. 1, arrows), digested, and analyzed by liquid chromatography (LC)/MS/MS, followed by SEQUEST database searching, which identified numerous proteins by matching the MS/MS spectra of identified peptides with the NCBI database. Identified proteins from each of the subcellular fractions only include those for which more than five matched peptides were found in concert with a sufficient correlation (summarized in Table 1). We emphasize that all the identified proteins are simply present in the excised immunopositive bands and that definitive identification of adducted amino acids within these proteins requires confirmation. Nonetheless, confidence that the immunoreactive proteins correspond to adducted proteins is provided by several additional key observations noted below.
Fig. 1.
Western blot analysis of MGHQ-treated Eker carrier rats cytosol (A), microsome (B), mitochondria (C), plasma membrane (D), and nuclei (E) from kidney OSOM segment. Eker rats (Tsc-2Ek/+) were treated with MGHQ (400 μmol/kg, i.v.) or PBS vehicle. Kidney OSOM tissue was isolated and collected in the isolation buffer, and the subcellular fractionation was carried out immediately after as described under Materials and Methods. CB, Coomassie Blue stain; WB, Western blot immunostain. Each subcellular fraction of treated or control OSOM proteins (100 μg/lane) was separated on duplicate 10% SDS-PAGE reducing gels. One gel was transferred to nitrocellulose and probed with anti-2-BrHQ-NAC antibodies followed by ECL. The other gel was stained with Coomassie Blue. Molecular mass markers are indicated on the far left lanes. Arrows indicate immunoreactive bands and their corresponding CB-stained gel band that was excised for LC/MS/MS analysis.
TABLE 1.
Summary of proteins present in QT-immunoreactive bands identified by LC/MS/MS analysis of kidney fractions
| Protein | Accession | Number of Peptides | Subcellular Fractions* | % Amino Acid Coveragea | Molecular Mass | pI | Protein Description and Function |
|---|---|---|---|---|---|---|---|
| Calbindin | P07171 | 10 | C | 42.7 | 29,863 | 4.71 | Calbindin, vitamin D-dependent calcium-binding protein |
| Carbonic anhydrase 2 | P27139 | 5 | C | 23.9 | 28,983 | 6.88 | Carbonic anhydrase II, Carbonate dehydratase II |
| Glutamate—cysteine ligase catalytic subunit | P19468 | 11 | C | 20 | 72,488 | 5.41 | γ-Glutamylcysteine synthetase, rate-limiting step enzyme for GSH biosynthesis |
| Glutamate—cysteine ligase regulatory subunit | P48508 | 5 | C | 23.7 | 30,548 | 5.36 | γ-Glutamylcysteine synthetase, rate-limiting step enzyme for de novo GSH biosynthesis |
| Heat shock protein HSP 90-α | P07901 | 10 | C | 18.3 | 84,657 | 4.93 | Heat shock protein HSP 86, tumor-specific transplantation 86-kDa antigen |
| Heat shock protein HSP 90-β | P34058 | 13 | C | 21.7 | 83,185 | 4.97 | Heat shock protein HSP 90-β, molecular chaperone has ATPase activity |
| Malate dehydrogenase, cytoplasmic | P14152 | 9 | C | 29.7 | 36,346 | 6.16 | Malate dehydrogenase, cytosolic, a tricarboxylic acid cycle enzyme |
| Transketolase | P50137 | 11 | C | 14.6 | 67,631 | 7.22 | Transketolase, involved in sugar metabolism, synthesis of ribose |
| Triosephosphate isomerase | P48500 | 6 | C | 28.2 | 26,790 | 7.06 | Triosephosphate isomerase TIM |
| Heat shock-related 70-kDa protein 2 | P14659 | 5 | C, Mic | 6.6 | 69,529 | 5.51 | Heat shock-related 70-kDa protein 2, HSP 70.2, HST |
| Heat shock cognate 71-kDa protein | P63018 | 11 | C, Mic | 18.7 | 70,871 | 5.37 | Heat shock cognate 71-kDa protein, constitutively synthesized chaperone protein |
| Actin, cytoplasmic 2 | P63259 | 16 | C, Mic, Mito, Nu, Pl | 41.3 | 41,793 | 5.31 | Cytoplasmic 2, γ-actin, cytoskeletal protein, mediator of internal cell motility |
| Elongation factor 1-α1 | P62630 | 8 | C, Mic, Pl | 16.9 | 50,114 | 9.10 | Elongation factor 1-α chain, promotes ribosome protein synthesis |
| Catalase | P04762 | 10 | C, Mito | 25.2 | 59,757 | 7.18 | Catalase, decomposes hydrogen peroxide to water and oxygen |
| α-Enolase | P04764 | 13 | C, Mito | 30.9 | 46,985 | 6.16 | α-Enolase, 2 phospho-d-glycerate hydrolyase, glycolytic enzyme in glycolysis |
| Glutathione S-transferase α-1 | P00502 | 10 | C, Mito, Pl | 29.4 | 25,402 | 8.87 | GSH S-transferase GT41A, class α, conjugates GSH to varieties of xenobiotics |
| Fructose-bisophosphate aldolase B | P00884 | 17 | C, Mito, Pl | 38 | 39,487 | 8.67 | Fructose-bisphosphate aldolase B, glycolytic enzyme involved in glycolysis |
| Serum albumin [Precursor] | P02770 | 14 | C, Pl | 22.5 | 68,719 | 5.80 | Rat serum albumin, good binding protein, regulates blood colloidal osmotic pressure |
| Clathrin heavy chain 1 | P11442 | 7 | Mic | 5.9 | 191,597 | 5.50 | Clathrin heavy chain |
| Aminopeptidase N | P15684 | 14 | Mito | 16.6 | 109,317 | 5.30 | Aminopeptidase N, microsomal, membrane protein cleaves N-amino acid off a peptide |
| Atrophin-1 | P54258 | 5 | Mito | 6.2 | 113,054 | 8.97 | Na/K transporting ATPase α 1 chain, Na pump |
| ATP synthase subunit, α, mitochondrial [Precursor] | P15999 | 18 | Mito | 34.6 | 58,826 | 8.28 | ATP synthase α chain, regulatory subunit, it produces ATP in mitochondria |
| Moesin | P26041 | 5 | Mito | 6.8 | 67,636 | 6.24 | Moesin, membrane organizing extension spike protein |
| Radixin | Q5PQK5 | 4 | Mito | 5.3 | 68,452 | 5.95 | Radixin |
| ATP synthase subunit β, mitochondrial [Precursor] | P10719 | 17 | Mito | 38.8 | 26,354 | 4.95 | ATP synthase β chain, catalytic subunit, produces ATP in mitochondria |
| Ezrin | P26040 | 8 | Mito | 10.9 | 69,215 | 5.83 | Ezrin p81, cytovillin, may connect cytoskeletal structures to plasma membrane |
C, cytosol; Mic, microsome; Mito, mitochondria; Nu, nucleus; Pl, plasma membrane.
The highest number of peptides matched.
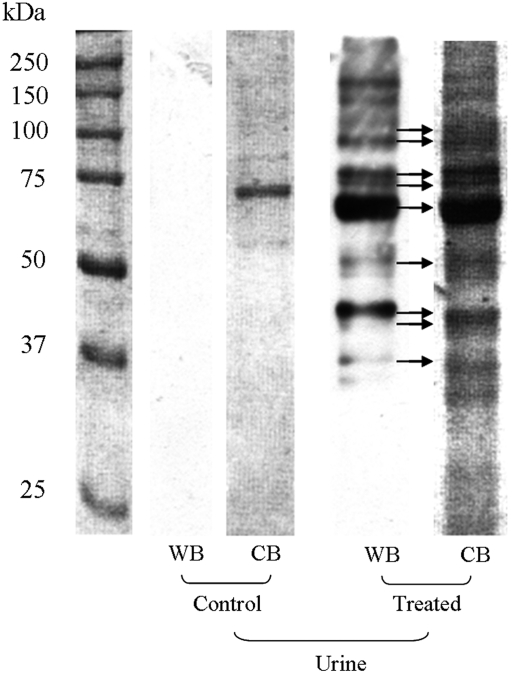
Western Blot Analysis Followed by In-Gel Digestion and LC/MS/MS Protein Identification for Urine. Fewer immunopositive proteins were visualized in the MGHQ-treated rat urine compared with kidney fractions (Fig. 2). Proteins excreted in urine from control rats were immunonegative. Proteins identified from immunoreactive bands from Western analysis of urine samples are summarized in Table 2. Urine samples contained fewer immunopositive bands than the cytosolic, microsomal, and mitochondrial fractions from the OSOM of kidneys from MGHQ-treated animals, probably because of protease-mediated degradation in situ. However, it is noteworthy that immunopositive proteins identified in urine were almost always present in the subcellular fractions of the OSOM, with the exception of argininosuccinate synthase citrulline-aspartate, a urea cycle arginine biosynthetic pathway enzyme; glutathione synthetase, second step of glutathione biosynthesis; kynurenine/α-aminoadipate aminotransferase; and rat transferrin, an iron-binding serum transport glycoprotein (Table 2). It is possible that these immunopositive proteins arise via their interaction with reactive urinary metabolites of MGHQ within the urinary tract and/or bladder, and that adducts on these particular proteins hinder protease-mediated digestion.
Fig. 2.
Western blot analysis of urinary proteins from MGHQ-treated Eker rats. Eker rats (Tsc-2EK/+) were treated with MGHQ (400 μmol/kg, i.v.) or PBS vehicle. Proteins from 2-h urine samples (200 μg/lane) were separated on 10% SDS-PAGE reducing gels. The gel was transferred to nitrocellulose and probed with anti-2-BrHQ-NAC antibodies followed by ECL. Molecular mass markers are indicated on the far left lane. Arrows indicate immunoreactive bands and their corresponding CB-stained gel band that was excised for LC/MS/MS analysis.
TABLE 2.
Summary of QT immunoreactive proteins in urine
| Protein | Accession | Number of Peptides | Molecular Mass | pI | Protein Description and Function |
|---|---|---|---|---|---|
| Actin, cytoplasmic 2 | P63259 | 6 | 41,963 | 5.31 | α-Actin, aortic smooth muscle, cytoskeleton protein, mediator of internal cell motility |
| Serum albumin [Precursor] | P02770 | 10 | 68,719 | 5.80 | Rat serum albumin, good binding protein, regulates blood colloidal osmotic pressure |
| Aminopeptidase N | P15684 | 11 | 109,430 | 5.30 | Aminopeptidase N, microsomal, membrane protein cleaves N-amino acid off a peptide |
| Argininosuccinate synthase | P09034 | 3 | 46,448 | 7.63 | Argininosuccinate synthase citrulline-aspartate, urea cycle arginine biosynthetic pathway enzyme |
| Glutathione synthetase | P46413 | 3 | 52,326 | 5.48 | GSH synthetase, second-step GSH biosynthesis enzyme |
| Glutamate-cysteine ligase catalytic | P19468 | 6 | 72,601 | 5.41 | Glutamate-cysteine ligase catalytic subunit, heavy chain, first-step glutathione biosynthesis enzyme |
| Kynureninase | P70712 | 8 | 47,735 | 6.19 | Kynurenine/α-aminoadipate aminotransferase, involves in NAD cofactors biosynthesis |
| Serotransferrin | P12346 | 7 | 75,840 | 6.99 | Rat transferrin, iron binding serum transport glycoprotein |
Common Protein Targets of Quinol-Thioethers and Other Electrophiles. Contrary to established dogma, adduction of proteins by reactive electrophiles is not a random event, but rather specific proteins seem to be targeted. These protein targets may differ between different electrophiles but also will have a varying degree of overlap. Chemical structure, reactivity, and ability to localize within the various intracellular compartments are several characteristics that govern which proteins are targeted by a given electrophile. The QT-targeted proteins identified in Tables 1 and 2 were compared with the target protein database (Hanzlik et al., 2007), which catalogs proteins adducted with reactive intermediates, and 15 of the 29 proteins were identified as targets of other electrophiles (Table 3). The majority of overlapping proteins were identified as targets in the liver, where the majority of reactive metabolites are generated. The chemicals with shared protein targets with QT exhibited both diverse (mycophenolic acid and atrazine) and similar (naphthalene, benzene, bromobenzene, and acetaminophen) chemical structures.
TABLE 3.
QT immunoreactive proteins targeted by other electrophiles
| Protein | Accession | Chemical | Tissue | Species |
|---|---|---|---|---|
| Actin, cytoplasmic 2 | P63259 | BHT benzene | Lung, bone marrow | Mouse (Meier et al., 2005) |
| Mouse (Williams et al., 2002) | ||||
| Aminopeptidase N | P15684 | Diclofenac | Intestine | Rat (Ware et al., 1998) |
| ATP synthase subunit α, mitochondrial [Precursor] | P15999 | Acetaminophen | Liver | Mouse (Qiu et al., 1998) |
| Benzene | Liver | Mouse (Williams et al., 2002) | ||
| ATP synthase subunit β, mitochondrial [Precursor] | P10719 | Naphthalene | Lung | Mouse (Lin et al., 2005) |
| Benzene | Liver | Mouse (Williams et al., 2002) | ||
| Carbonic anhydrase 2 | P27139 | Mycophenolic acid | Kidney | Rat (Asif et al., 2007) |
| Catalase | P04762 | Thiobenzamide | Liver | Rat (Ikehata et al., 2008) |
| Teucrin A | Liver | Rat (Druckova et al., 2007) | ||
| Glutathione S-transferase α-1 | P00502 | Bromobenzene | Liver | Rat (Koen et al., 2006) |
| Mycophenolic acid | Kidney | Rat (Asif et al., 2007) | ||
| Heat shock cognate 71-kDa protein | P63018 | Bromobenzene | Liver | Rat (Koen et al., 2007) |
| Mycophenolic acid | Kidney | Rat (Asif et al., 2007) | ||
| Thiobenzamide | Liver | Rat (Ikehata et al., 2008) | ||
| Heat shock protein HSP 90 | P34058 | Acetaminophen | Liver | Mouse (Qiu et al., 1998) |
| 3-Hydroxyacetanilide | Liver | Mouse (Qiu et al., 2001) | ||
| BHT | Lung | Mouse (Meier et al., 2005) | ||
| Mycophenolic acid | Kidney | Rat (Asif et al., 2007) | ||
| Teucrin A | Liver | Rat (Druckova et al., 2007) | ||
| Malate dehydrogenase, cytoplasmic | P14152 | Thiobenzamide | Liver | Rat (Ikehata et al., 2008) |
| Mycophenolic acid | Kidney | Rat (Asif et al., 2007) | ||
| Radixin | Q5PQK5 | BHT | Liver | Rat (Reed and Thompson, 1997) |
| Triosephosphate isomerase | P48500 | BHT | Lung | Mouse (Meier et al., 2005) |
| Bromobenzene | Liver | Rat (Koen et al., 2007) | ||
| Mycophenolic acid | Kidney | Rat (Asif et al., 2007) |
Identification of Electrophile-Binding Motifs. We searched for common binding motifs within MGHQ-adducted proteins (Person et al., 2003, 2005; Fisher et al., 2007). The cytochrome c sequences GGKHKTG (23–29) and GIKKK (84–88) were identified by LC/MS/MS as binding sites for BQ (Person et al., 2003), MGHQ (Person et al., 2005), and 2-(N-acetylcystein-S-yl)-hydroquinone (Fisher et al., 2007). The lysine residues are thought to participate in forming a cyclized diquinone adduct (Person et al., 2003). Therefore, we searched the primary sequences (obtained via the Swiss-Prot database) of the identified proteins for the presence of basic amino acid run-ons. It is interesting to note that all 30 adducted proteins possess lysine-rich sequences. In particular, adducted proteins contain lysine residues either flanking a potentially nucleophilic amino acid (KXK) or contain two lysine residues preceded or followed by a nucleophilic amino acid (XKK or KKX). The presence of 1, 2, 3, 4 to 6, 7 to 9, 10 to 20, and >20 lysine-rich sequences were found in one, five, four, eight, seven, three, and two proteins, respectively. Adducted proteins that contain more than nine XKK or KKX sequences are identified in Table 4, and of the six such proteins identified with an abundance of the EBM, each one possesses an elevated lysine content (9.7%) compared with the average protein lysine content within the entire proteome (5.5%).
TABLE 4.
Occurrence of EBM (KXK, KK) in target proteins of QT adduction
| Heat Shock Protein 90-α 11% Lysine P07901 | Heat Shock Protein 90-β 10% Lysine P34058 | Moesin 11% Lysine P26041 | Radixin 11% Lysine Q5PQK5 | Clathrin Heavy Chain 16% Lysine P11442 | Ezrin 9% Lysine P26040 |
|---|---|---|---|---|---|
| 205 EIVKKHSQ 212 | 200 EVVKKHSQ 207 | 60 KLNKKVTA 67 | 60 KLNKKVTQ 67 | 242 PFPKKAVD 249 | 60 KLDKKVSA 67 |
| 267 EEEKKDGD 274 | 262 GKDKKKKT 269 | 208 IKNKKGSE 215 | 68 QDVKKENP 75 | 503 LYAKKVGY 510 | 208 IKNKKGTD 215 |
| 272 DGDKKKKK 279 | 263 KDKKKKTK 270 | 250 FNDKKFVI 257 | 208 IKNKKGTE 215 | 1159 MARKKARE 1166 | 250 FNDKKFVI 257 |
| 273 GDKKKKKK 280 | 264 DKKKKTKK 271 | 259 PIDKKAPD 266 | 250 FNDKKFVI 257 | 1526 ELCKKDSL 1533 | 259 PIDKKAPD 266 |
| 274 DKKKKKKI 281 | 267 KKTKKIKE 274 | 324 ENEKKKRE 331 | 259 PIDKKAPD 266 | 93 IEMKSKMKA 101 | 324 ETEKKRRE 331 |
| 275 KKKKKKIK 282 | 344 FENKKKKN 351 | 325 NEKKKREL 332 | 324 ENEKKKRE 331 | 95 MKSKMKAHT 103 | 206 FEIKNKKGT 214 |
| 276 KKKKKIKE 283 | 345 ENKKKKNN 352 | 356 EQTKKAQQ 363 | 325 NEKKKREI 332 | 158 TDAKQKWLL 166 | 354 YEQKTKRAE 362 |
| 354 ENRKKKNN 361 | 346 NKKKKNNI 353 | 408 RDQKKTQE 415 | 432 EEAKKKKE 439 | 1323 LYSKFKPQK 1331 | 455 DLVKTKEEL 463 |
| 355 NRKKKNNI 362 | 407 NIVKKCLE 414 | 433 MARKKKES 440 | 433 EAKKKKEE 440 | 1438 YFSKVKQLP 1446 | 561 GRDKYKTLR 569 |
| 416 NLVKKCLE 423 | 424 ENYKKFYE 431 | 434 ARKKKESE 441 | 434 AKKKKEEE 441 | ||
| 433 ENYKKFYE 440 | 547 EEEKKKME 554 | 534 DESKKTAN 541 | 522 ERVKKQLQ 529 | ||
| 455 QNRKKLSE 462 | 548 EEKKKMEE 555 | 206 FSIKNKKGS 214 | 540 DETKKTQN 547 | ||
| 556 EEEKKKQE 563 | 570 ILDKKVEK 577 | 324 ENEKKKREL 332 | 206 FEIKNKKGT 214 | ||
| 557 EEKKKQEE 564 | 620 MMAKKHLE 627 | 332 LAEKEKEKI 340 | 324 ENEKKKREI 332 | ||
| 562 QEEKKTKF 569 | 244 DEEKPKIED 252 | 334 EKEKEKIER 342 | 332 IAEKEKERI 340 | ||
| 579 ILEKKVEK 586 | 260 DSGKDKKKK 268 | 347 LMEKLKQIE 355 | 432 EEAKKKKEE 440 | ||
| 629 MAAKKHLE 636 | 262 GKDKKKKTK 270 | 433 MARKKKESE 441 | 433 EAKKKKEEE 441 | ||
| 242 EEEKEKEEK 250 | 263 KDKKKKTKK 271 | 552 GRDKYKTLR 560 | 455 DLEKTKEEL 463 | ||
| 272 DGDKKKKKK 280 | 265 KKKKTKKIK 273 | 558 GRDKYKTLR 566 | |||
| 273 GDKKKKKKI 281 | 268 KTKKIKEKY 276 | ||||
| 274 DKKKKKKIK 282 | 270 KKIKEKYID 278 | ||||
| 275 KKKKKKIKE 283 | 281 ELNKTKPIW 289 | ||||
| 277 KKKKIKEKY 285 | 344 FENKKKKNN 352 | ||||
| 279 KKIKEKYID 287 | 345 ENKKKKNNI 353 | ||||
| 290 ELNKTKPIW 298 | 547 EEEKKKMEE 555 | ||||
| 354 ENRKKKNNI 362 | 554 EESKAKFEN 562 | ||||
| 556 EEEKKKQEE 564 | |||||
| 563 EEKKTKFEN 571 |
Discussion
In the present study, we identified 30 proteins from Western blots expressing QT-immunoreactive bands. These proteins were identified either from kidney samples (Table 1; n = 26) or urine (Table 2; n = 8) of MGHQ-treated Eker rats. Confidence that proteins identified from the immunoreactive bands also contain MGHQ-derived adducts includes the following observations: 1) of the eight immunoreactive proteins identified in urine, four were also identified in kidney samples; 2) of the 30 proteins identified in the present study, a remarkable 43% (n = 13) are targets of other electrophiles (Table 3); 3) of the 13 QT immunoreactive protein targets (Table 3) that are also targets of other electrophiles, nine are targets of chemicals that give rise to either quinones, quinoneimines, and/or quinone methides; and 4) all 30 adducted proteins possess lysine-rich sequences previously identified as specific targets of QTs. Given the size and diversity of the proteome, the probability that such findings are random in nature is highly unlikely. Finally, although it remains possible that some of the proteins identified in the immunoreactive bands represent false-positives, several of the proteins identified herein have been shown, unequivocally, to contain QT-adducted peptides (M. T. Labenski, A. A. Fisher, J. D. Chapman, G. Tsaprailis, T. J. Monks, and S. S. Lau, unpublished observations).
Comparing the Coomassie-stained gel with the corresponding Western analysis (Figs. 1 and 2) revealed that only a select few cellular proteins were immunoreactive after exposure to MGHQ. The basis for this selectivity is unclear, but certain features probably predispose certain proteins and certain motifs within proteins to the adduction required to yield an immunoreactive protein (see below). The immunoreactive proteins (Tables 1 and 2) were subsequently compared with the target protein database (Hanzlik et al., 2007), which catalogs protein adducts from a structurally diverse array of chemicals that either directly adduct proteins or do so via the generation of reactive metabolites. Of the 30 proteins identified from immunopositive Western blots, an extraordinarily high fraction (43%) are targets of other electrophiles (Table 3). Therefore, the remaining 17 proteins represent novel targets.
Our data, in combination with the database compiled from the literature, support the existence of EBMs within proteins that are selectively adducted by reactive electrophiles. It is also likely that specific electrophiles (e.g., quinones, aldehydes, and episulfonium ions) target their own spectrum of EBMs based on unique postadduction chemistry and the stability/instability of the final adducts. However, it also seems that a single protein probably expresses multiple EBMs, hence the accumulating evidence that some proteins are targets of structurally diverse electrophiles. With respect to the reactive metabolites derived from MGHQ, QTs selectively target 1) proteins with a high lysine (basic amino acid) content, and specifically 2) proteins that contain lysine residues either flanking a potentially nucleophilic amino acid (KXK) or containing two lysine residues preceded or followed by a nucleophilic amino acid (XKK or KKX). It is interesting to note that, of the 13 QT immunoreactive protein targets (Table 3) that are also targets of other electrophiles, nine are targets of chemicals (acetaminophen, benzene, 2,6-di-tert-butyl-4-methylphenol, bromobenzene, 3-hydroxyacetanilide, and naphthalene) that give rise to quinones, quinoneimines, and/or quinone methides. The quinone moiety is a common feature of many drugs and is found in many natural products. Moreover, several endogenous compounds give rise to quinones, including β-estradiol and dopamine. For example, oxidation of dopamine generates the corresponding ortho-quinone, which binds to α-synuclein, preventing the protofibril to fibril conversion (Conway et al., 2001). This has direct relevance to the pathogenesis of Parkinson's disease, during which dopaminergic neurons within the substantia nigra accumulate fibrillar Lewy bodies composed primarily of α-synuclein. Thus, quinone adduct formation from dopamine provides a mechanistic explanation for the dopaminergic selectivity of α-synuclein-associated neurotoxicity in Parkinson's disease. It is noteworthy that wild-type α-synuclein does not contain cysteine, and we would predict that adduct formation is likely to occur on lysines and/or other basic amino acids. Indeed, α-synuclein is rich in lysines (10.7%) and contains several KXK, XKK, or KKX motifs.
Of the four proteins in Table 3 (aminopeptidase N, carbonic anhydrase, catalase, and cytoplasmic malate dehydrogenase) that had not been previously identified as targets of quinones, we would predict that they contain not only the KXK, XKK, or KKX motif but also other EBMs, including nucleophilic cysteines. Indeed, even cysteine adduction seems to be context (motif?)-dependent. For example, metallothionein (MT) contains no aromatic residues, and 20 of the 61 or 62 amino acid residues are cysteines. Regardless of this plethora of nucleophilic cysteine thiols in MT, selective binding precedes random alkylation because 90% of melphalan and 80% of chlorambucil react initially with Cys-34 and Cys-49 of MT (Yu et al., 1995; Zaia et al., 1996). These two cysteine residues cochelate the same metal ion in the α-domain and are in close proximity in the folded protein. In contrast, alkylation by mechlorethamine occurs predominantly in the carboxyl domain of MTs, with one molecule of mechlorethamine covalently cross-linking two cysteine residues (Antoine et al., 1998). Molecular dynamics simulation studies indicated that sites proximal to the sulfhydryl groups of Cys-33 and Cys-48 were the most favorable for complexing the aziridinium forms of chlorambucil, melphalan, and mechlorethamine (Szilagyi and Fenselau, 2000). Thus, selectivity of adduction is governed by both the recipient nucleophile and the chemistry of the adducting electrophile. Likewise, Cys-239 in tubulin is preferentially alkylated by anticancer drugs, including the colchicines, Vinca alkaloids, rhizoxin/maytansine, and taxanes (Jordan et al., 1998; Shan et al., 1999; Kavallaris et al., 2001; Casini et al., 2002; Scozzafava et al., 2002), and carbonic anhydrase III, which contains five cysteine residues, is selectively alkylated at Cys-186 by acrylonitrile (Nerland et al., 2003).
Cysteine thiols are clearly common targets of reactive electrophilic metabolites and endogenous electrophiles, including lipid-derived α,β-unsaturated aldehydes, such as 4-hydroxynonenal (4-HNE) and 4-oxononenal, and reactive oxygen species. Although in most cases the toxicological significance of protein adducts remains uncertain, physiological concentrations of either 4-HNE or 4-oxononenal cause the cross-linking of bovine brain tubulin and an inability of tubulin to polymerize (Stewart et al., 2007). 4-HNE also reduces extracellular signal-regulated kinase 1/2 phosphorylation, causing a loss of activity and in nuclear localization. It is interesting to note that the loss of extracellular signal-regulated kinase activity is caused by a single 4-HNE modification on histidine 178 (Sampey et al., 2007). However, inactivation of glyceraldehyde 3-phosphate dehydrogenase by acrylonitrile (Campian et al., 2002), as well as inactivation of glutathione S-transferase-Z by dichloroacetic acid (Anderson et al., 2002) and maleylacetone and fumaryl-acetone (Lantum et al., 2002), occurs via adduction of cysteine residues. In some instances, the covalent binding of anticancer drugs to cellular proteins represents a detoxication process. For example, alkylating mustard anticancer drugs, including melphalan, mechlorethamine, and chlorambucil, form covalent adducts with MTs (Kotsonis and Klaassen, 1979; Endresen et al., 1983; Yu et al., 1995; Zaia et al., 1996; Antoine et al., 1998), which contributes to the inactivation of these drugs.
In summary, although the concentration and reactivity of macromolecular nucleophiles will certainly influence the pattern of macromolecular alkylation, only those macromolecules with accessible (surface exposed?) nucleophilic groups (such as protein sulfhydryls and amines) will become adducted. Motifs contained within each target protein most likely contribute to their attraction to QTs. For example, cyclic QT reaction products on cytochrome c are preferentially formed at sites where multiple basic residues exist in a conformationally flexible region, whereas noncyclic products bind to a broad spectrum of generally solvent accessible lysine and histidine nucleophiles (Person et al., 2005). Thus, the microenvironment at the site of adduction (solvent accessibility and local pKa) probably governs both the initial specificity and the structure of the final adduct. Postadduction chemistry may subsequently alter the nature of the final adduct (Fisher et al., 2007). The relative selectivity of adduction might also depend on the exact location of the protein within the cell, and as emphasized by Gillette et al. (1984), the concentration and half-life (reactivity?) of the reactive metabolite. The critical nature of the target will be defined by the function and accessibility of the protein adducted, not by the selectivity of the reactive electrophile. Approaches such as those described herein will assist in clarifying which combination of parameters contribute to the susceptibility of specific proteins to chemical adduction, and the functional consequences thereof.
Acknowledgments
We thank Dr. George Tsaprailis (Director of the Arizona Protein Consortium) and Sean Davey (Informatics Specialist at the Bioinformatics Facility Core of the Southwest Environmental Health Sciences Center) for producing custom software Motif_Hunter.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant R01-GM070890]; and the National Institutes of Health National Institute of Environmental Health Sciences [Grants T32-ES07091, P30-ES006694, P30-ES007784].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.026211.
ABBREVIATIONS: MS, mass spectrometry; HPLC, high-performance liquid chromatography; BQ, 1,4-benzoquinone; QT, quinol-thioether; EBM, electrophile-binding motif; MGHQ, 2-(glutathion-S-yl)-hydroquinone; 2-BrHQ-NAC, 2-bromo-6-(N-acetylcystein-S-yl)hydroquinone; ECL, enhanced chemiluminescence; PBS, phosphate-buffered saline; γ-GT, γ-glutamyltranspeptidase; OSOM, outer stripe of the outer medulla; PAGE, polyacrylamide gel electrophoresis; MS/MS, tandem mass spectrometry; amu, atomic mass unit; NCBI, National Center for Biotechnology Information; KLH, keyhole-limpet hemocyanin; LC, liquid chromatography; MT, metallothionein; 4-HNE, 4-hydroxynonenal.
References
- Anderson WB, Liebler DC, Board PG, and Anders MW (2002) Mass spectral characterization of dichloroacetic acid-modified human glutathione transferase zeta. Chem Res Toxicol 15 1387–1397. [DOI] [PubMed] [Google Scholar]
- Antoine M, Fabris D, and Fenselau C (1998) Covalent sequestration of the nitrogen mustard mechlorethamine by metallothionein. Drug Metab Dispos 26 921–926. [PubMed] [Google Scholar]
- Asif AR, Armstrong VW, Voland A, Wieland E, Oellerich M, and Shipkova M (2007) Proteins identified as targets of the acyl glucuronide metabolite of mycophenolic acid in kidney tissue from mycophenolate mofetil treated rats. Biochimie 89 393–402. [DOI] [PubMed] [Google Scholar]
- Campian EC, Cai J, and Benz FW (2002) Acrylonitrile irreversibly inactivates glyceraldehyde-3-phosphate dehydrogenase by alkylating the catalytically active cysteine 149. Chem Biol Interact 140 279–291. [DOI] [PubMed] [Google Scholar]
- Casini A, Scozzafava A, and Supuran CT (2002) Cysteine-modifying agents: a possible approach for effective anticancer and antiviral drugs. Environ Health Perspect 110 (Suppl 5): 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KA, Rochet JC, Bieganski RM, and Lansbury PT Jr (2001) Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science 294 1346–1349. [DOI] [PubMed] [Google Scholar]
- Craig R and Beavis RC (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom 17 2310–2316. [DOI] [PubMed] [Google Scholar]
- Druckova A, Mernaugh RL, Ham AJ, and Marnett LJ (2007) Identification of the protein targets of the reactive metabolite of teucrin A in vivo in the rat. Chem Res Toxicol 20 1393–1408. [DOI] [PubMed] [Google Scholar]
- Endresen L, Bakka A, and Rugstad HE (1983) Increased resistance to chlorambucil in cultured cells with a high concentration of cytoplasmic metallothionein. Cancer Res 43 2918–2926. [PubMed] [Google Scholar]
- Fisher AA, Labenski MT, Malladi S, Gokhale V, Bowen ME, Milleron RS, Bratton SB, Monks TJ, and Lau SS (2007) Quinone electrophiles selectively adduct “electrophile binding motifs” within cytochrome c. Biochemistry 46 11090–11100. [DOI] [PubMed] [Google Scholar]
- Gillette JR, Lau SS, and Monks TJ (1984) Intra- and extra-cellular formation of metabolites from chemically reactive species. Biochem Soc Trans 12 4–7. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Cai H, Johnson WW, and Parikh A (2001) Reactive intermediates in biological systems: what have we learned and where are we going? Adv Exp Med Biol 500 639–650. [DOI] [PubMed] [Google Scholar]
- Hanzlik RP, Koen YM, Theertham B, Dong Y, and Fang J (2007) The reactive metabolite target protein database (TPDB)–a web-accessible resource. BMC Bioinformatics 8 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehata K, Duzhak TG, Galeva NA, Ji T, Koen YM, and Hanzlik RP (2008) Protein targets of reactive metabolites of thiobenzamide in rat liver in vivo. Chem Res Toxicol 21 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Hadfield JA, Lawrence NJ, and McGown AT (1998) Tubulin as a target for anticancer drugs: agents which interact with the mitotic spindle. Med Res Rev 18 259–296. [DOI] [PubMed] [Google Scholar]
- Kavallaris M, Verrills NM, and Hill BT (2001) Anticancer therapy with novel tubulin-interacting drugs. Drug Resist Updat 4 392–401. [DOI] [PubMed] [Google Scholar]
- Kleiner HE, Jones TW, Monks TJ, and Lau SS (1998a) Immunochemical analysis of quinol-thioether-derived covalent protein adducts in rodent species sensitive and resistant to quinol-thioether-mediated nephrotoxicity. Chem Res Toxicol 11 1291–1300. [DOI] [PubMed] [Google Scholar]
- Kleiner HE, Rivera MI, Pumford NR, Monks TJ, and Lau SS (1998b) Immunochemical detection of quinol–thioether-derived protein adducts. Chem Res Toxicol 11 1283–1290. [DOI] [PubMed] [Google Scholar]
- Koen YM, Gogichaeva NV, Alterman MA, and Hanzlik RP (2007) A proteomic analysis of bromobenzene reactive metabolite targets in rat liver cytosol in vivo. Chem Res Toxicol 20 511–519. [DOI] [PubMed] [Google Scholar]
- Koen YM, Yue W, Galeva NA, Williams TD, and Hanzlik RP (2006) Site-specific arylation of rat glutathione S-transferase A1 and A2 by bromobenzene metabolites in vivo. Chem Res Toxicol 19 1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsonis FN and Klaassen CD (1979) Increase in hepatic metallothionein in rats treated with alkylating agents. Toxicol Appl Pharmacol 51 19–27. [DOI] [PubMed] [Google Scholar]
- Lantum HB, Liebler DC, Board PG, and Anders MW (2002) Alkylation and inactivation of human glutathione transferase zeta (hGSTZ1-1) by maleylacetone and fumarylacetone. Chem Res Toxicol 15 707–716. [DOI] [PubMed] [Google Scholar]
- Lau SS, Hill BA, Highet RJ, and Monks TJ (1988) Sequential oxidation and glutathione addition to 1,4-benzoquinone: correlation of toxicity with increased glutathione substitution. Mol Pharmacol 34 829–836. [PubMed] [Google Scholar]
- Lau SS, Monks TJ, Everitt JI, Kleymenova E, and Walker CL (2001) Carcinogenicity of a nephrotoxic metabolite of the “nongenotoxic” carcinogen hydroquinone. Chem Res Toxicol 14 25–33. [DOI] [PubMed] [Google Scholar]
- Lin CY, Isbell MA, Morin D, Boland BC, Salemi MR, Jewell WT, Weir AJ, Fanucchi MV, Baker GL, Plopper CG, et al. (2005) Characterization of a structurally intact in situ lung model and comparison of naphthalene protein adducts generated in this model vs lung microsomes. Chem Res Toxicol 18 802–813. [DOI] [PubMed] [Google Scholar]
- Meier BW, Gomez JD, Zhou A, and Thompson JA (2005) Immunochemical and proteomic analysis of covalent adducts formed by quinone methide tumor promoters in mouse lung epithelial cell lines. Chem Res Toxicol 18 1575–1585. [DOI] [PubMed] [Google Scholar]
- Nerland DE, Cai J, and Benz FW (2003) Selective covalent binding of acrylonitrile to Cys 186 in rat liver carbonic anhydrase III in vivo. Chem Res Toxicol 16 583–589. [DOI] [PubMed] [Google Scholar]
- Person MD, Monks TJ, and Lau SS (2003) An integrated approach to identifying chemically induced posttranslational modifications using comparative MALDI-MS and targeted HPLC-ESI-MS/MS. Chem Res Toxicol 16 598–608. [DOI] [PubMed] [Google Scholar]
- Person MD, Mason DE, Liebler DC, Monks TJ, and Lau SS (2005) Alkylation of cytochrome c by (glutathion-S-yl)-1,4-benzoquinone and iodoacetamide demonstrates compound-dependent site specificity. Chem Res Toxicol 18 41–50. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Benet LZ, and Burlingame AL (2001) Identification of hepatic protein targets of the reactive metabolites of the non-hepatotoxic regioisomer of acetaminophen, 3′-hydroxyacetanilide, in the mouse in vivo using two-dimensional gel electrophoresis and mass spectrometry. Adv Exp Med Biol 500 663–673. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Benet LZ, and Burlingame AL (1998) Identification of the hepatic protein targets of reactive metabolites of acetaminophen in vivo in mice using two-dimensional gel electrophoresis and mass spectrometry. J Biol Chem 273 17940–17953. [DOI] [PubMed] [Google Scholar]
- Reed M and Thompson DC (1997) Immunochemical visualization and identification of rat liver proteins adducted by 2,6-di-tert-butyl-4-methylphenol (BHT). Chem Res Toxicol 10 1109–1117. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, and Ferrara P (1992) In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem 203 173–179. [DOI] [PubMed] [Google Scholar]
- Sampey BP, Carbone DL, Doorn JA, Drechsel DA, and Petersen DR (2007) 4-Hydroxy-2-nonenal adduction of extracellular signal-regulated kinase (Erk) and the inhibition of hepatocyte Erk-Est–like protein-1-activating protein-1 signal transduction. Mol Pharmacol 71 871–883. [DOI] [PubMed] [Google Scholar]
- Scozzafava A, Casini A, and Supuran CT (2002) Targeting cysteine residues of biomolecules: new approaches for the design of antiviral and anticancer drugs. Curr Med Chem 9 1167–1185. [DOI] [PubMed] [Google Scholar]
- Shan B, Medina JC, Santha E, Frankmoelle WP, Chou TC, Learned RM, Narbut MR, Stott D, Wu P, Jaen JC, et al. (1999) Selective, covalent modification of beta-tubulin residue Cys-239 by T138067, an antitumor agent with in vivo efficacy against multidrug-resistant tumors. Proc Natl Acad Sci U S A 96 5686–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BJ, Doorn JA, and Petersen DR (2007) Residue-specific adduction of tubulin by 4-hydroxynonenal and 4-oxononenal causes cross-linking and inhibits polymerization. Chem Res Toxicol 20 1111–1119. [DOI] [PubMed] [Google Scholar]
- Szilágyi Z and Fenselau C (2000) Molecular dynamics simulation of metallothionein-drug complexes. Drug Metab Dispos 28 174–179. [PubMed] [Google Scholar]
- Ware JA, Graf ML, Martin BM, Lustberg LR, and Pohl LR (1998) Immunochemical detection and identification of protein adducts of diclofenac in the small intestine of rats: possible role in allergic reactions. Chem Res Toxicol 11 164–171. [DOI] [PubMed] [Google Scholar]
- Williams KE, Carver TA, Miranda JJ, Kautiainen A, Vogel JS, Dingley K, Baldwin MA, Turteltaub KW, and Burlingame AL (2002) Attomole detection of in vivo protein targets of benzene in mice: evidence for a highly reactive metabolite. Mol Cell Proteomics 1 885–895. [DOI] [PubMed] [Google Scholar]
- Yu X, Wu Z, and Fenselau C (1995) Covalent sequestration of melphalan by metallothionein and selective alkylation of cysteines. Biochemistry 34 3377–3385. [DOI] [PubMed] [Google Scholar]
- Zaia J, Jiang L, Han MS, Tabb JR, Wu Z, Fabris D, and Fenselau C (1996) A binding site for chlorambucil on metallothionein. Biochemistry 35 2830–2835. [DOI] [PubMed] [Google Scholar]