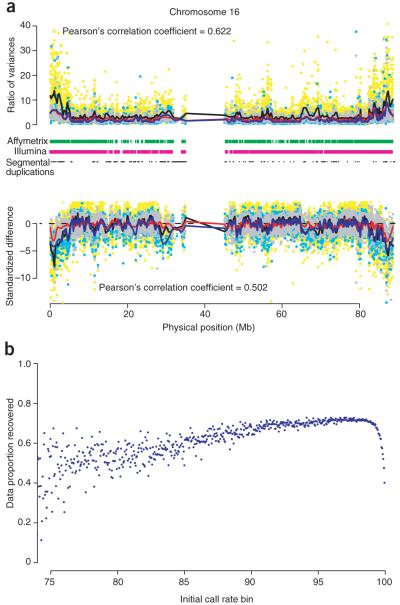
Figure 1.
Missingness and imputation of amplified DNA on chromosome 16. (a) The relative performance of amplified DNA to genomic DNA, as quantified by the ratio of hybridization strengths and the standardized difference in mean hybridization strength. Each plot shows data from three comparisons of amplified DNA to genomic DNA. Affymetrix: TB (cyan dots and blue lines); Affymetrix: OBC-58C (gray dots and red lines); Illumina: ML-58C (yellow dots and black lines). Lines below the upper plots indicate regions where SNPs on the platform have call rates < 95.0%. The dashes in black indicate regions of segmental duplications. (b) The expected proportion of missing genotypes which can be recovered using the program IMPUTE6 as a function of the initial call rate. Initial SNP call rates have been partitioned into 0.01 bins with each bin’s data proportion recovery averaged across the number of SNPs.