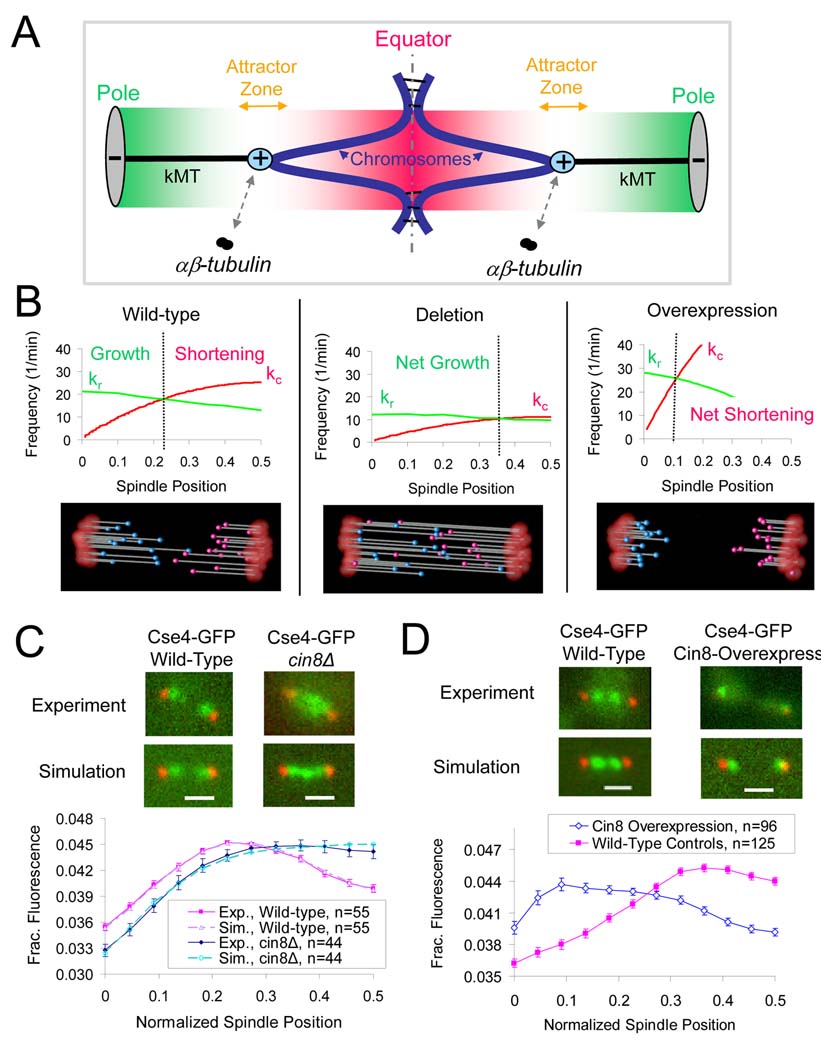
Figure 1. Cin8p organizes metaphase yeast kinetochores in a manner consistent with length-dependent suppression of net kMT plus-end assembly.
(A) A spatial gradient in net kMT plus-end assembly mediates kinetochore congression in yeast. Kinetochores (cyan) congress to attractor zones (yellow arrows) on either side of the spindle equator (dotted line) during yeast metaphase via the plus-end assembly dynamics of kMTs (black). The kMT plus-end assembly dynamics are spatially regulated such that plus-end assembly is favored near the poles (grey) when kMTs are relatively short (favorable assembly zone shown as green gradient), and suppressed near the spindle equator (dotted line) when kMTs are relatively long (assembly suppression zone shown as red gradient). (B) A computational model of yeast metaphase kMT plus end dynamics predicts that deletion of the length-dependent promoter of kMT disassembly will result in longer kMTs and disorganized kinetochores. Conversely, overexpression will result in shorter kMTs and focused kinetochore clusters. All model parameters as in Table S2. (C) CIN8 deletion results in kinetochore disorganization and the net shifting of kinetochores towards the spindle equator, suggesting that kMTs are on average longer than they are in wild-type cells (red, Spc29-CFP pole marker; green, Cse4-GFP kinetochore marker) (scale bar 1000 nm, error bars, s.e.m.) (D) Cin8p overexpression results in clusters of kinetochores near the SPBs, suggesting that kMTs are shorter than in wild-type cells.