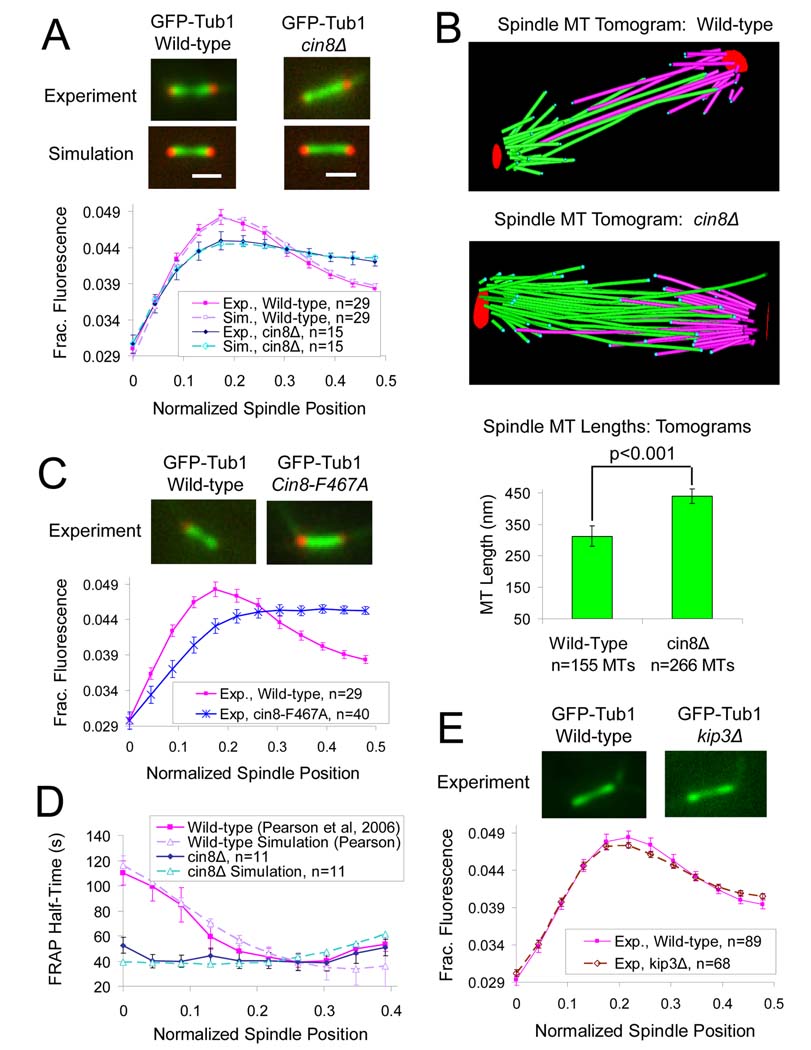
Figure 2. Cin8p promotes kMT disassembly.
(A) CIN8 deletion results in flattening of the GFP-tubulin fluorescence distribution and shifting of fluorescence towards the spindle equator, suggesting that kMT lengths are increased in the mutant. (Red, Spc29 CFP SPB marker; Green, GFP-Tub1 MT marker) (scale bar 1000 nm, error bars, s.e.m.) All model parameters as in Table S2. (B) Cryo-electron tomography reveals increased mean MT length and number in cin8Δ spindles (n=5 spindles, mean spindle length =1387 nm), relative to wild-type spindles (n=4 spindles, mean spindle length=1265 nm). (C) Similar to the CIN8 deletion mutant, the motor domain mutant, cin8-F467A, which has reduced affinity for MTs, results in flattening of the GFP-tubulin fluorescence distribution and shifting of fluorescence towards the spindle equator. (D) CIN8 deletion eliminates the characteristic gradient in GFP-tubulin FRAP recovery half-time, consistent with disruption of the gradient in net kMT assembly. (E) In contrast to the cin8Δ mutants, deletion of the kinesin-8 depolymerase KIP3 does not significantly perturb kinetochore microtubule (kMT) organization.