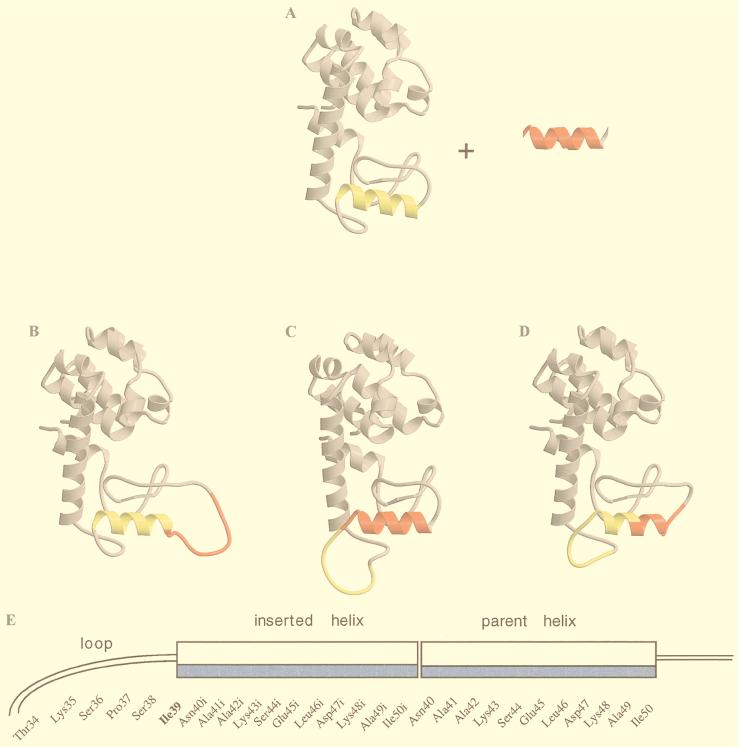
Figure 1.
Model structures illustrating some possible structural responses to sequence duplication. The original helix sequence is highlighted in yellow and the sequence of the insert in red. (A) Wild-type T4 lysozyme. (B) The inserted sequence is looped out at the amino terminus of the parent helix. (C) Looping out at the carboxyl terminus of the parent helix. (D) A helix similar to that in WT* lysozyme is formed in part by the parent sequence and in part by the inserted sequence. Loop structures would be formed at each end of the helix. (E) Sketch showing the sequence of the inserted region (Asn-40i–Ile-50i) relative to the sequence and secondary structure of wild-type lysozyme. If the parent and the insert were to form a single, continuous α-helix, it would be amphipathic with the hydrophobic side shown shaded. To maintain this continuous hydrophobic surface, and also to ensure that the sequence included an exact 11-aa repeat, Leu-39 in the wild-type sequence was replaced by an isoleucine and is designated Ile-39. As a consequence the connection between the two helices is not interrupted by any residues that contribute to loops or other nonhelical structures in the native protein. Figure drawn with bobscript (19), molscript (20), and raster3d (21).