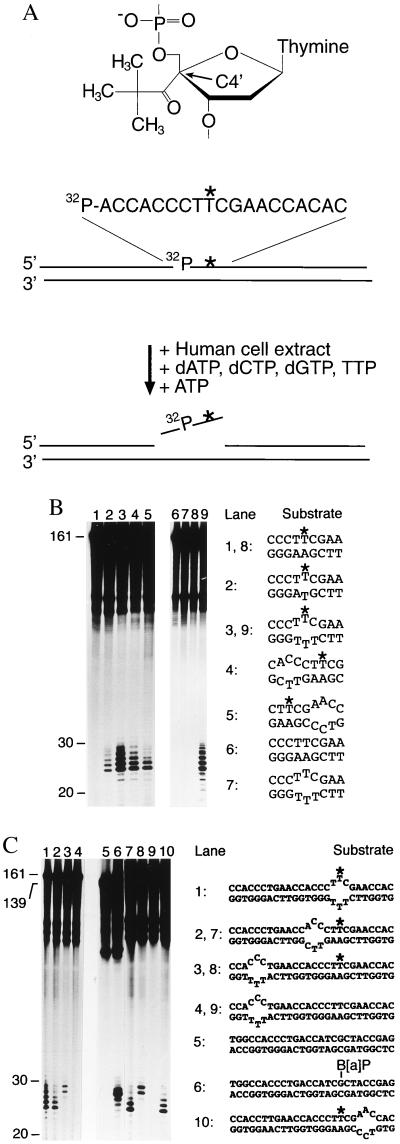
Figure 1.
Recruitment of human NER activity depends on defective hybridization. (A) Structure of the nondistorting C4′ pivaloyl adduct (Top), basic design of the 161-mer substrates (Middle) and oligonucleotide excision assay (Bottom). The C4′ pivaloyl modification is indicated by ∗, and a 32P-labeled residue is situated on the 5′ side of this lesion. (B) Determination of human NER activity in HeLa cell extract. After incubations of 40 min at 30°C, excision products were visualized by denaturing gel electrophoresis and autoradiography. All reactions contained the same amount of radioactive substrate (5 fmol), and oligonucleotide lengths were estimated by using appropriate markers. These incubations in cell extract also generate nonspecific degradation products, visible in the upper part of each lane, which do not interfere with detection of excised oligomers. The sequence environment in the central portion of each substrate is indicated, with the asterisks denoting the C4′ pivaloyl adduct. (C) Probing of NER activity using composite substrates. The central portion of each double-stranded fragment is outlined, again with ∗ denoting the C4′ modification. Lane 6 shows a control reaction with a (−)-cis-B[a]P-modified substrate of 139 bp (12).