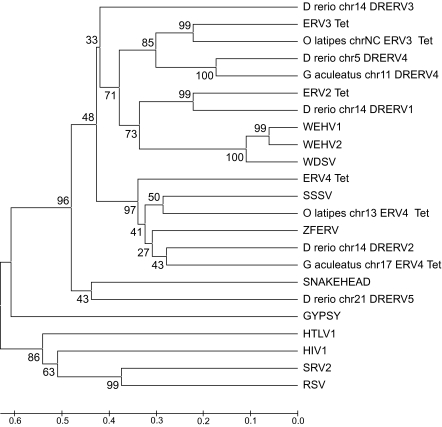
Figure 10.
Phylogenetic tree of fish retroviruses. The tree was constructed using RT amino acid sequences from 23 retroviruses (both endogenous and exogenous) and only includes RTs which have all six motifs of the OSM (see Methods). This tree was made using the MEGA 3.0 software (Kumar et al. 2004), using the UPGMA (Sneath et al. 1973) method with bootstrap values (3000 repetitions) (Felsenstein et al. 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl et al. 1985) and are in the units of the number of amino acid substitutions per site. Organism name, chromosome number, and then Retroid name label tree tips for those novel agents pulled out by a query. The retroviruses and accession numbers that are not included on the query host organism tree (Figure 3) are Walleye epidermal hyperplasia virus type 1 (WEHV1) (AF014792), Walleye epidermal hyperplasia virus type 2 (WEHV1) (AF014793), Atlantic salmon swim bladder sarcoma virus (SSSV) (DQ174103) and Rous sarcoma virus (RSV) (NC_001407). In addition to RSV, GYPSY, HTLV1, HIV1 and SRV2 (Figure 3) are included as non-fish-retrovirus out groups.