Table 1.
Structure statistics
| NOE restraints | |
| Total | 1,086 |
| Intraresidue | 548 |
| Sequential | 263 |
| Medium range | 87 |
| Long range | 188 |
| φ dihedral angle restraints | 26 |
| Hydrogen bond restraints | 30 |
| Deviation from experimental restraints | |
| Distance restraints, Å | 0.014 ± 0.003 |
| Dihedral restraints, deg | 0.17 ± 0.09 |
| Deviation from idealized covalent geometry | |
| Bonds, Å | 0.002 ± 0.000 |
| Angles, deg | 0.48 ± 0.01 |
| Impropers, deg | 0.359 ± 0.005 |
| Backbone rms deviation, Å | |
〈SA〉 to 〈 〉 residues A147-L226 〉 residues A147-L226 |
1.45 ± 0.16 |
〈SA〉 to 〈 〉 all β-strands 〉 all β-strands |
0.80 ± 0.08 |
〈SA〉 stands for the ensemble of 20 NMR structures and 〈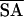 〉 is the average structure of the ensemble calculated by using x-plor. The parameter used to calculate the van der Waals (vdw) repulsion energy was 0.75 rather than 0.80 (47).
〉 is the average structure of the ensemble calculated by using x-plor. The parameter used to calculate the van der Waals (vdw) repulsion energy was 0.75 rather than 0.80 (47).
