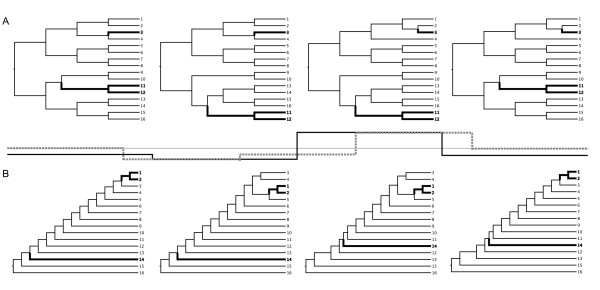
Figure 3.
Phylogenetic simulation of recombinant sequences. Sequence data sets were simulated according to (a) symmetric and (b) asymmetric tree topologies. Each tree represents a single partition of 1000 nucleotides. In the symmetric combination, this results in a single breakpoint mosaic pattern for taxon 3 and a shared dual breakpoint mosaic pattern for taxon 11 and 12. In the asymmetric combination, taxon 1 and 2 share a relative recent recombination event with two breakpoints, while taxon 14 is a descendent of an older single breakpoint recombination event. In between both sets of trees, the relative rate profiles are plotted relative to 1 (thin line): the black line pattern represents rate variation correlated with the breakpoint positions (0.75, 0.50, 2.00 and 0.75), while the grey dashed line represents rate variation for partitions of 800 bp (1.0, 0.50, 0.75, 2.00 and 0.75).