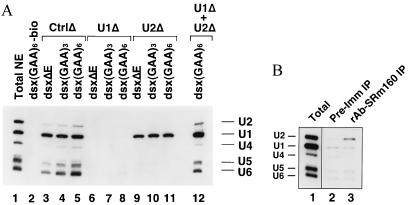
Figure 3.
Interactions between SRm160/300 and snRNPs in the assembly of dsx splicing complexes. (A) U1 snRNP binds to the dsx pre-mRNA independently of the ESE, whereas the ESE and U1 snRNP are required for the assembly of U2, U4/U6, and U5 snRNPs on the dsx pre-mRNA. Biotinylated dsx pre-mRNAs were incubated in mock-depleted or snRNP-depleted splicing reaction mixtures for 40 min before affinity selection on streptavidin-agarose. RNA recovered from the beads was separated on a denaturing 10% polyacrylamide gel and analyzed by Northern hybridization using riboprobes specific for the five spliceosomal snRNAs. Lanes 3–11 contain RNA recovered after affinity selection with biotinylated dsx pre-mRNAs, and lane 2 contains RNA recovered after a control selection performed in the presence of a nonbiotinylated dsx(GAA)6 pre-mRNA. Selections were performed using the dsx pre-mRNA with no enhancer sequence (lanes 3, 6, 9), with the (GAA)3 enhancer (lanes 4, 7, 10), or with the (GAA)6 enhancer (lanes 5, 8, 11, 12) from splicing reaction mixtures containing “mock-depleted” extract (CtrlΔ, lanes 2–5), U1 snRNP-depleted extract (U1Δ, lanes 6–8), or U2 snRNP-depleted extract (U2Δ, lanes 9–11). The selection in lane 12 was performed from a splicing reaction mixture containing an equal mixture of the U1 and U2 snRNP-depleted extracts. Lane 1 contains RNA recovered directly from nuclear extract, representing approximately 3% of the amount of extract used in each selection. (B) A subpopulation of U2 snRNP associates with SRm160/300 in the absence of exogenous pre-mRNA. RNA recovered after immunoprecipitation with rAb-SRm160 (lane 3) or a corresponding preimmune serum (lane 2) from HeLa nuclear extract (lane 1) was analyzed as in A. The amount of nuclear extract represented in lane 1 corresponds to approximately 5% of the amount used for each immunoprecipitation.