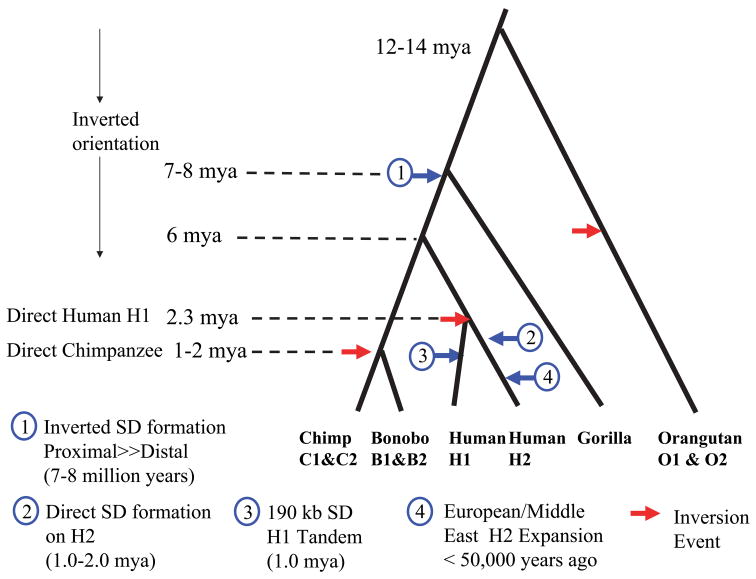
Figure 5. Evolutionary Model: Inversion “toggling”, SD formation and disease susceptibility.
We estimated the evolutionary age of various duplication/gene conversion and rearrangement events by establishing a local molecular clock for single nucleotide substitution (Supplementary Table 4) and superimposed these estimates over a generally accepted hominoid phylogeny 45. Due to uncertainty in the chimp-human divergence, timing of events should only be considered an approximation. We propose that the ancestral CRHR1-MAPT region was inverted but has toggled to an H1 orientation multiple times within the evolution of different great-ape/human lineages (red arrows). Large blocks (≥100 kbp) of inverted segmental duplications were formed in the common ancestor of chimpanzee and human, further predisposing the region to recurrent inversion. An inversion of the predominant H2 allele created the H1 allele ~2.3 million years ago within the human lineage. Subsequently, larger blocks of directly oriented SDs emerged within the H2 lineage predisposing it to microdeletion and disease. As a result of this negative selection against the H2, the H1 haplotype rose in frequency and became the predominant allele in all human populations with a subsequent CNP tandem duplication occurring on some haplotypes. In the out-of-Africa European founding population, however, the H2 allele resurged in frequency due to a partial selective sweep or a population bottleneck in the founding population.