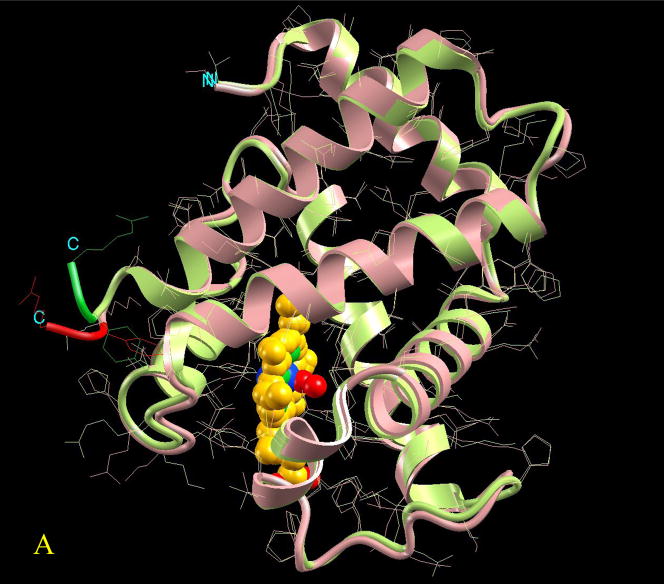
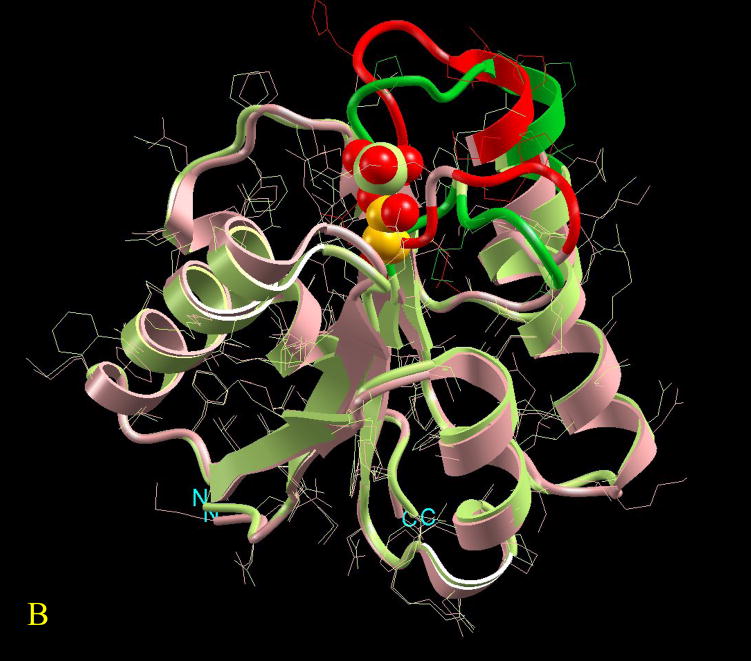
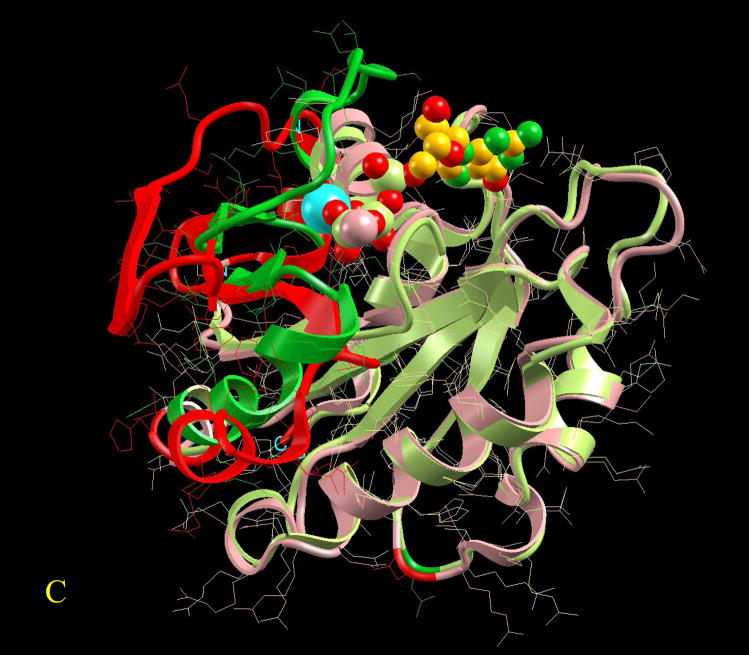
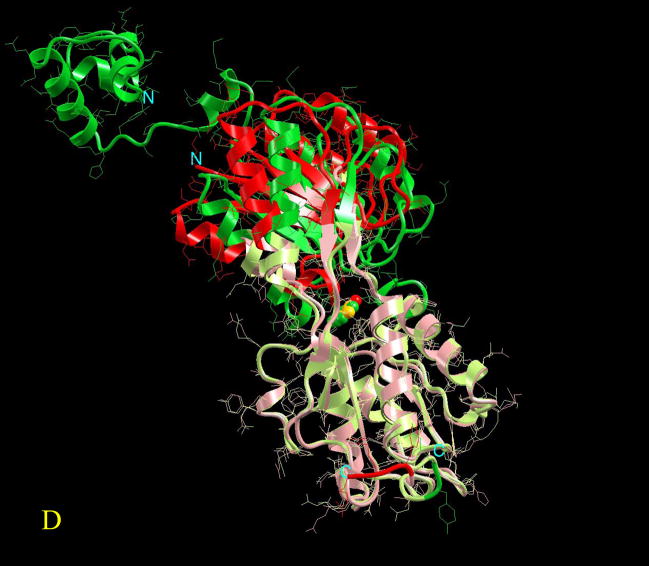
Figure 3.
Visualization of the overall conformational changes in allosteric proteins. Four pairs of known inactive and active allosteric protein structures from the Protein Data Bank are illustrated. In each pair, the protein backbone trace is represented by solid ribbon and the side-chains are drawn as thin lines. The effector molecule binding (or covalent modification) to the active allosteric protein is shown in space-fill (atom color codes are Carbon yellow, Nitrogen green, Oxygen red, Phosphorus light green, Sulfur pink, Magnesium cyan, and Iron blue). The ligand is located at the allosteric site. However, even if present in the PDB file of the inactive protein, it is not shown here for clarity. The superposition is based on matched residues with the distance between superimposed Cα atoms <= 2.0 Å. The scaffold of the matched residues is colored pink and light green, respectively for the inactive and active allosteric proteins. The conformational changes (unmatched residues) are highlighted in red and green, respectively for the inactive and active allosteric proteins. Here four pictures are illustrated as examples of conformational changes in allostery: (A) hemoglobin (PDB codes: 2hhb A and 1hho A); this case has been classified in the no-change category, (B) fixJ (1dbw A; 1d5w A): classified as subtle (C) arf6 (1e0s A; 1hfv A): classified minor, (D) purR (1dbq A; 1wet A): classified domain-movement.