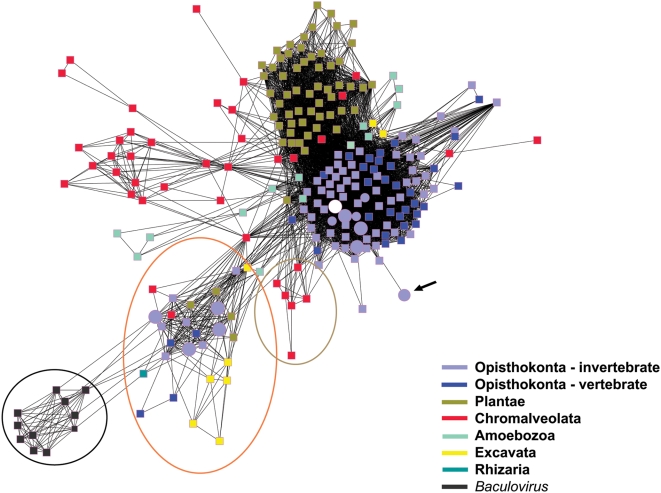
Figure 12. Sequence relationships among the cathepsin L enzymes.
Clustering of cathepsin L sequences accords with the organization of the kingdoms of life into six super kingdoms [45]: Opisthokonta (light purple and dark blue nodes), Plantae (green), Chromalveolata (red), Amoebozoa (aquamarine), Rhizaria (single turquoise node) and Excavata (yellow). Circular nodes represent groups of cathepsin L sequences from platyhelminths and those that are enlarged identify cathepsin L sequences specific to Trematoda. Square nodes depict cathepsin L sequences from other groups. The white circle represents the sequence from SmCL3 and its S. japonicum ortholog, SjCL3, which are found among the opisthokont, invertebrate metazoan cathepsin L genes. The large circular node more distant from the main opisthokont cluster corresponds to an inactive cathepsin L ortholog from S. japonicum (black arrow). The network view identifies a diverse group of cathepsin L sequences making up a cluster enriched in cathepsin L subtypes cathepsins F and W (encircled in orange) that includes sequences from S. mansoni, S. japonicum, O. viverrini, C. sinensis, P. westermani and M. yokogawai. Encircled in green are sequences exclusive to the cathepsin H subtype of cysteine cathepsins L. Finally, a cluster composed solely of baculovirus cathepsin L genes (within the black circle) is least connected to all other sequences in the network.