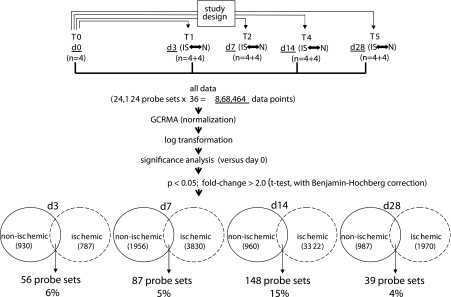
Fig. 7.
GeneChip data analysis scheme used to identify the kinetics of differentially expressed genes in nonischemic and ischemic wounds. Image acquisition and processing was performed using GCOS (GeneChip operating software, Affymetrix). GC-RMA was applied for data normalization. ArrayAssist v. 5.1 software was used to identify significant (P < 0.05; false discovery rate corrected) differentially expressed genes in nonischemic wound tissue compared with skin (0 h). The list of wound-sensitive genes thus obtained was queried for differential expression under ischemic vs. nonischemic conditions. The numbers of such wound-responsive ischemia-sensitive transcripts are indicated at the bottom together with a percentage value that represents the fraction of the total number of wound-responsive transcripts that were ischemia-sensitive for the corresponding time point after wounding.