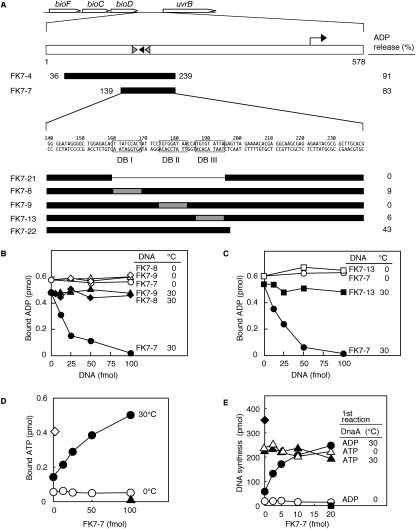
Figure 1.
Structure and in vitro activity of DARS1. (A) Identification of DARS1. Open arrows indicate the coding regions. The right-angled arrow indicates the promoter. Black- and gray-filled arrowheads represent DnaA boxes that completely match the 9-mer consensus and that differ from the consensus by a single base, respectively. Numbers above the sequence of FK7-7 indicate the position from the stop codon of bioD. FK7-8, FK7-9, and FK7-13 contain nonsense sequence (AACTATATC; gray-filled square) (Schaper and Messer 1995) instead of DnaA box I (DB I), DnaA box II (DB II), and DnaA box III (DB III), respectively. [3H]ADP-DnaA (2 pmol) was incubated for 15 min at 30°C in buffer containing 2 mM ATP in the presence (50 fmol) or absence of the indicated DNA fragment (filled bars). DNA-dependent release of ADP is shown for each fragment (ADP release %). (B,C) ADP-releasing activity of DARS1 mutants. The indicated amounts of FK7-7 (○, ●) or its derivatives FK7-8 (◇, ♦), FK7-9 (△, ▲), and FK7-13 (□, ■) were incubated at 0°C (unfilled symbols) or 30°C (filled symbols) for 15 min with [3H]ADP-DnaA (2 pmol). (D) Nucleotide-exchanging activity of DARS1. ADP-DnaA (2 pmol) was incubated for 15 min at 0°C (○) or 30°C (●) with 1.5 μM [α-32P]ATP and the indicated amounts of FK7-7. [α-32P]ATP-DnaA (2 pmol) was similarly incubated at 0°C (◇). (▲) DnaA was not included. (E) Reactivation of DnaA by DARS1 for replication initiation. ADP-DnaA (○, ●) and ATP-DnaA (△, ▲) (0.4 pmol) were incubated (first reaction) for 15 min at 0°C (○, ▵) or 30°C (●, ▲) with 2 mM ATP and the indicated amounts of FK7-7. The samples were then further incubated for 20 min at 30°C in a minichromosome replication system. (■) minichromosome was not included; (♦) DNA synthesis by 1 pmol of ATP-DnaA.