Abstract
Proteases are found in every cell, where they recognize and break down unneeded or abnormal polypeptides or peptide-based nutrients within or outside the cell. Genome sequence data can be used to compare proteolytic enzyme inventories of different organisms as they relate to physiological needs for protein modification and hydrolysis. In this review, we exploit genome sequence data to compare hyperthermophilic microorganisms from the euryarchaeotal genus Pyrococcus, the crenarchaeote Sulfolobus solfataricus, and the bacterium Thermotoga maritima. An overview of the proteases in these organisms is given based on those proteases that have been characterized and on putative proteases that have been identified from genomic sequences, but have yet to be characterized. The analysis revealed both similarities and differences in the mechanisms utilized for proteolysis by each of these hyperthermophiles and indicated how these mechanisms relate to proteolysis in less thermophilic cells and organisms.
Keywords: Archaea, Bacteria, protease, Pyrococcus, Sulfolobus solfataricus, Thermotoga maritima
Introduction
Proteases are critical to the maintenance of cellular function. They hydrolyze both external and internal nutrient sources and recognize and break down unneeded or abnormal polypeptides, the latter produced as a result of environmental stress, mutation or errors in biosynthetic processes (Tomoyasu et al. 2001). Cells have an array of proteases for processing proteins and polypeptides (e.g., Allison and Macfarlane 1990, Guedon et al. 2001) and for maintaining metabolic function under abnormal conditions (Arsene et al. 2000). Proteases range from simple monomeric hydrolases to complex, multi-subunit structures with molecular masses in the order of 1 MDa. Some proteases have a high functional and structural complexity, particularly those that are ATP-dependent, e.g., Lon, ClpXP, ClpYQ, FtsH and the proteasome (Gottesman 1996, Maupin-Furlow et al. 2000), but whether simple or complex, all proteases must be precisely regulated to avoid destruction of the cell’s metabolic machinery.
Here, we exploit genomic sequence data, in conjunction with physiological, biochemical and biophysical information, to investigate relationships among proteolytic inventories of hyperthermophilic microorganisms (capable of growth at 80 °C) of the euryarchaeotal genus Pyrococcus, the crenarchaeote Sulfolobus solfataricus and the bacterium Thermotoga maritima. Thermotoga maritima, initially isolated from Vulcano, Italy, has an optimum growth temperature of 80 °C and is a fermentative anaerobe that reduces sulfur facultatively and prefers simple and complex sugars as growth substrates (Huber et al. 1986). Sulfolobus solfataricus, initially isolated from a solfataras field near Naples, Italy (Zillig et al. 1980), is an aerobe that grows at acidic pHs as low as 2.0, with an optimal growth temperature of 80 °C (Brock et al. 1972). This archaeon grows chemolithotrophically by oxidizing metal cations (Fe2+) or sulfur as well as heterotrophically on simple sugars. Pyrococcus is a marine hyperthermophilic genus that includes species with optimal growth temperatures between 96 and 100 °C. Pyrococcus spp. have been isolated from deep-sea hydrothermal vent systems, such as those found along the North Fiji basin (P. abyssi, Erauso et al. 1993) and Okinawa Trough (P. horikoshii, Gonzalez et al. 1998), as well as from shallow marine environments, such as those found around Vulcano Island, Italy (P. furiosus, Fiala and Stetter 1986). Pyrococcus spp. are strict anaerobes that reduce sulfur facultatively and grow heterotrophically by fermentation of proteinaceous compounds and sometimes simple sugars and α-keto acids such as pyruvate.
Inferring protease inventory from genomic sequences
Initial efforts to assess the extent and variety of proteases in hyperthermophiles by biochemical methods significantly underestimated this biocatalytic feature. For example, Connaris et al. (1991) and Blumentals et al. (1990) reported that gelatin-based zymograms of cell-free extracts from P. furiosus revealed the presence of up to 13 clearing zones, some of which were later attributed to multiple versions of a single protease (Halio et al. 1996, Chang et al. 2001). Similar experiments with T. maritima revealed an even more limited set of proteases than observed in P. furiosus (Hicks et al. 1998). Genome sequence data, however, indicate that the proteolytic genotypes of these organisms are more expansive than can be inferred from zymogram analyses. Tables 1 and 2 show the confirmed and putative protease-related genes in the genomes of P. furiosus (http://comb5-156.umbi.umd.edu/genemate/), P. abyssi, P. horikoshii (Kawarabayasi et al. 1998), S. solfataricus (She et al. 2001)and T. maritima (Nelson et al. 1999). This information was also used to examine other hyperthermophilic and mesophilic organisms (for more information see http://www.che.ncsu.edu/extremophiles/). Protease-related genes were identified through BLAST analyses of completely sequenced prokaryotic genomes, the online database at the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.ad.jp/kegg/), and the AlignAce output files (http://arep.med.harvard.edu/microbial_motifs/). Putative protease/peptidases were analyzed for known motifs with ScanProsite (http://ca.expasy.org/tools/scnpsit1.html), the presence of signal peptides using SignalP (http://www.cbs.Dtu.dk/services/SignalP/) and genomic organization using the STRING analysis method (Snel et al. 2000), for possible protease-gene relationships. Homology was confirmed when amino acid sequence identities were at least 25% over 50% or more of the protein. Genomic analysis showed that Pyrococcus spp., S. solfataricus and T. maritima have numerous protease homologs to putative and confirmed proteases in other archaea and bacteria, although there are many differences among these organisms in protease inventory. This is true even when comparing the protease inventory of the three species within the genus Pyrococcus.
Table 1.
Protease inventory in Pyrococcus species, Sulfolobus solfataricus and Thermotoga maritima.
| Pyrococcus furiosus | Pyrococcus abyssi | Pyrococcus horikoshii | Solfolobus solfataricus | Thermatoga maritima | |||||
| Locus | Function | Locus | Function | Locus | Function | Locus | Function | Locus | Function |
| ATP-dependent proteases | ATP-dependent proteases | ATP-dependent proteases | ATP-dependent proteases | ATP-dependent proteases | |||||
| pf_481472 | ATP-dependent LA (Lon)1 | PAB1313 | ATP-dependent LA (Lon)1 | PH0452 | ATP-dependent LA (Lon)1 | SSO0278 | Proteasome beta subunit | TM1633 | ATP-dependent protease LA (Lon) |
| pf_119316 | ATP-dependent regulatory subunit | PAB2233 | ATP-dependent regulatory subunit | PH0201 | ATP-dependent regulatory subunit | SSO0738 | Proteasome alpha subunit | TM1869 | ATP-dependent protease LA, putative |
| pf_169930 | Proteasome beta subunit (PsmB-2) | PAB2199 | Proteasome beta subunit (PsmB-2) | PH0245 | Proteasome beta subunit (PsmB-2) | SSO0766 | Proteasome beta subunit | TM0198 | ATP-dependent Clp protease, ATPase (clpC-1) |
| pf_1318448 | Proteasome beta subunit (PsmB-1) | PAB1867 | Proteasome beta subunit (PsmB-1) | PH1402 | Proteasome beta subunit (PsmB-1) | SSO0271 | 26S proteosome regulatory subunit | TM1391 | ATP-dependent Clp protease, ATPase (clpC-3) |
| pf_1467425 | Proteasome alpha subunit | PAB0417 | Proteasome alpha subunit | PH1553 | Proteasome alpha subunit | TM0146 | ATP-dependent Clp protease, ATPase (clpX) | ||
| ATP-independent proteases | TM0873 | ATP-dependent Clp protease, ATPase (clpC-2) frameshift | |||||||
| ATP-independent proteases | ATP-independent proteases | ATP-independent proteases | SSO1886 | Thermopsin precursor, probable | TM0695 | ATP-dependent Clp protease, proteolytic subunit (clpP) | |||
| pf_301754 | Pyrolysin | PAB1252 | Pyrolysin-like | PH0310 | Pyrolysin-like | SSO2037 | Thermopsin precursor, probable | TM0521 | Heat-shock protein HslV (clpQ) proteolytic subunit |
| pf_251719 | Periplasmic serine protease, putative | PAB2437 | Periplasmic serine protease, putative | PH0282 | Periplasmic serine protease, putative | SSO2194 | Thermopsin precursor related protein | TM0522 | Heat-shock protein HslU (clpY) ATPase |
| pf_176949 | Metalloprotease | PAB2195 | Protease, putative | PH0256 | Protease, putative | SSO2551 | Serine protease, subtilase family, putative | TM0580 | Cell division protein FtsH |
| pf_401364 | Metalloprotease | PAB1063 | Metalloprotease | PH0351 | Metalloprotease | SSO1141 | Protease related protein | ||
| pf_472643 | Metalloprotease | PAB1319 | Metalloprotease | PH1569 | Hypothetical protease IV | SSO2045 | Protease | ATP-independent proteases | |
| pf_699579 | Subtisilin-like protease | PAB1960 | Hypothetical protease IV | PH1704 | Intracellular protease I (PfpI) | SSO2098 | Tricorn protease | TM0747 | Carboxy-terminal protease |
| pf_1479679 | Protease IV | PAB0311 | Intracellular protease I (PfpI) | PH0979 | Hydrogenase maturation protease (hyc I) | SSO1067 | Intracellular protease (PfpI) | TM0516 | Clostripain-related protein |
| pf_1553191 | Alkaline serine protease | PAB0576 | Hydrogenase maturation protease (hyc I) | PH0630 | Hypothetical neutral protease | SSO1759 | Protease, putative | TM0643 | Clostripain-related protein |
| pf_1757236 | Putative protease | PAB0555 | CAAX Prenyl protease | PH0585 | Hypothetical neutral protease | SSO2015 | Protease, putative | TM1589 | Clostripain-related protein |
| pf_1599202 | Intracellular protease I (PfpI) | SSO2612 | HtrA-like Serine protease (periplasmic) | TM1823 | FtsH protease activity modulator HflC (hflC) | ||||
| pf_637303 | Hydrogenase maturation protease (hyc I) | Peptidases | Peptidases | SSO0464 | Hypothetical protein, put. protease | TM1822 | FtsH protease activity modulator HflK (hflK) | ||
| pf_754933 | Hypothetical protein | PAB1320 | Carboxypeptidase I174 | PH0465 | Carboxypeptidase I | SSO0087 | Hypothetical protein, put. metalloprotease | TM0571 | Heat-shock serine protease, periplasmic (htrA) |
| pf_1136394 | Conserved hypothetical protein (bacteriocin homolog) | PAB1418 | Protein similar to acylaminoacyl peptidase | PH0594 | Protein similar to acylaminoacyl peptidase | TM1346 | Processing protease, putative | ||
| PAB1159 | O-sialoglycoprotein endopeptidase (gcp-1) | PH1987 | O-sialoglycoprotein endopeptidase (gcp-1) | Peptidases | TM0785 | Protease, bacteriocin | |||
| Peptidases | PAB1047 | O-sialoglycoprotein endopeptidase (gcp-2) | PH0445 | O-sialoglycoprotein endopeptidase (gcp-2) | SSO3115 | Tricorn Protease interacting F factor F1 | TM0145 | Secreted metalloendopeptidase Gcp, putative | |
| pf_471039 | Carboxypeptidase I | PAB1434 | Methionine aminopeptidase (MAP) | PH0628 | Methionine aminopeptidase (MAP) | SSO2154 | Tricorn Protease interacting F factor F2 | TM0409 | Conserved hypothetical, probable secreted proteinase |
| pf_334012 | Protein similar to acylaminoacyl peptidase | PAB2178 | Proline-dipeptidase related protein | PH1902 | Proline-dipeptidase related protein | SSO2675 | Tricorn Protease interacting F factor F3 | TM0890 | Conserved hypothetical, probable membrane metalloprotease |
| pf_181525 | O-sialoglycoprotein endopeptidase (gcp-1) | PAB2178 | X-Pro dipeptidase | PH1902 | Putative proline-dipeptidase | SSO0010 | X-pro aminopeptidase | TM1704 | Hypothetical protein (homology to protease maturation) |
| pf_488067 | O-sialoglycoprotein endopeptidase (gcp-2) | PAB0762 | Prolyl endopeptidase | PH1262 | Prolyl endopeptidase | SSO0363 | Prolidase | TM0916 | Periplasmic serine protease, putative |
| pf_561253 | Methionine aminopeptidase (MAP) | PAB2388 | Membrane dipeptidase | PH1026 | Membrane dipeptidase | SSO0433 | O-sialoglycoprotein endopeptidase | ||
| pf_707740 | Proline-dipeptidase related protein | PAB0045 | D-aminopeptidase | PH0078 | D-aminopeptidase | SSO0434 | O-sialoglycoprotein endopeptidase | Peptidases | |
| pf_743892 | Putative proline-dipeptidase | PAB1637 | XAA-Pro dipeptidase (proline dipeptidase) | PH1149 | XAA-Pro dipeptidase (proline dipeptidase) | SSO1355 | Thermostable carboxypeptidase (cpsA-1) | TM0963 | Oligoendopeptidase, putative |
| pf_803543 | Prolyl endopeptidase | PAB1419 | Pyroglutamyl-peptidase I | PH0596 | Pyroglutamyl-peptidase I | SSO1952 | Thermostable carboxypeptidase (cpsA-2) | TM0847 | Conserved hypothetical protein |
| pf_848157 | Membrane dipeptidase | PAB2375 | Aminopeptidase from M42 family | PH0519 | Aminopeptidase from M42 family | SSO3105 | Thermostable carboxypeptidase 1 | TM0445 | Conserved hypothetical protein |
| pf_1775248 | D-aminopeptidase | PAB0437 | Aminopeptidase from M42 family | PH1527 | Aminopeptidase from M42 family | SSO1465 | Pyrrolidone carboxylate peptidase (pcp) | TM1572 | Signal peptidase I, putative |
| pf_1898123 | Putative aminopeptidase | PAB2096 | Aminopeptidase from M42 family | PH1821 | Aminopeptidase from M42 family | SSO1607 | Pyrrolidone carboxylate peptidase, put. (pcp-like) | TM1696 | Type IV prepilin peptidase |
| pf_1902688 | Putative aminopeptidase | PAB0723 | Acetylornithine deacetylase (ArgE)/peptidase | PH1153 | Acetylornithine deacetylase (ArgE)/peptidase | SSO1864 | Microsomal dipeptidase | TM0129 | Carboxypeptidase G2, putative |
| pf_1906416 | Putative aminopeptidase | PAB0581 | Succinyl-diaminopimelate desuccinylase/peptidase | PH1288 | Succinyl-diaminopimelate desuccinylase/peptidase | SSO0098 | Methionine aminoppetidase 2 | TM0874 | Conserved hypothetical protein |
| pf_1262501 | XAA-Pro dipeptidase (proline dipeptidase) | PAB0966 | Signal sequence peptidase I (SEC11) | PH0590 | Signal sequence peptidase I (SEC11) | SSO2088 | Peptidase, putative | TM0463 | Lipoprotein signal peptidase |
| pf_1224347 | Pyroglutamyl-peptidase I | PAB0869 | Peptidase, putative | PH0737 | Peptidase, putative | SSO2181 | Peptidase related protein | TM0772 | Conserved hypothetical |
| pf_383398 | Endoglucanase/peptidase | PAB1781 | X-Pro aminopeptidase pepQ-3 | PH0974 | Hypothetical dipeptidase | TM1048 | Endoglucanase (peptidase) | ||
| pf_1444497 | Endoglucanase (celM)/peptidase | TM1049 | Endoglucanase (peptidase) | ||||||
| pf_1717470 | Endo-1,4-beta-glucanase/peptidase | TM1050 | Endoglucanase (peptidase) | ||||||
| pf_1131554 | Acetylornithine deacetylase (ArgE)/peptidase | TM0042 | Aminopeptidase P, putative | ||||||
| pf_1890964 | Succinyl-diaminopimelate desuccinylase/peptidase | TM0365 | Aminopeptidase, putative | ||||||
| pf_329163 | Signal sequence peptidase I (SEC11) | TM1713 | Proline dipeptidase, putative | ||||||
| pf_679507 | Hypothetical protein | TM1478 | Methionine aminopeptidase (map) | ||||||
1The Lon protease in Pyrococcus lacks the ATP-binding domain, but was placed in the ATP-dependent protease section because there is no experimental evidence aside from the sequence to suggest otherwise.
Table 2.
Protease-related genes in selected microorganisms.
| Species | ATP-dependent proteases | ATP-independent proteases | Peptidases | Total |
| Thermotoga maritima | 9 | 14 | 16 | 39 |
| Pyrococcus furiosus | 5 | 13 | 22 | 40 |
| Pyrococcus abyssi | 5 | 9 | 20 | 34 |
| Pyrococcus horikoshii | 5 | 9 | 20 | 34 |
| Archaeoglobus fulgidus | 4 | 7 | 11 | 22 |
| Sulfolobus solfataricus | 4 | 13 | 16 | 33 |
| Escherichia coli | 13 | 16 | 31 | 60 |
The ATP-dependent proteases in hyperthermophiles
All prokaryotic genomes sequenced to date, including hyperthermophiles, indicate the presence of several ATP-dependent proteases, although there is some variation in the roster of these enzymes among organisms. These proteases are typically implicated in protein and peptide turnover, and stress response (for reviews see Gottesman 1996, 1999, Porankiewicz et al. 1999, Schmidt et al. 1999). Although it is unclear how particular functional or abnormal proteins are selected for proteolytic processing by ATP-dependent proteases, it is a key issue in understanding cellular function.
The Lon protease
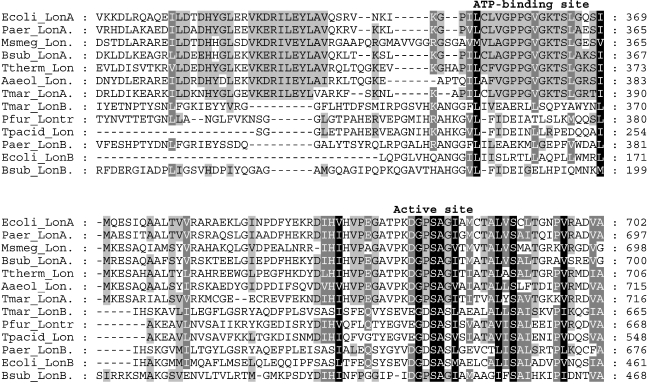
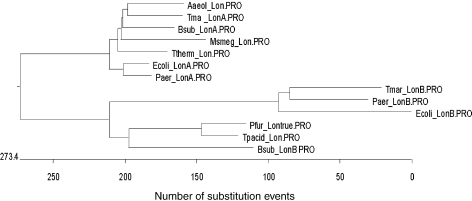
Lon protease is unique among ATP-dependent proteases in that it is not based on the assemblage of small subunits (i.e., 15–25 kDa) into stacked rings that interact with separate ATPases. Rather, Lon protease is a homotetramer, composed of 87-kDa subunits, each containing an active site serine and a single ATP-binding site (Maurizi 1992). Lon is ubiquitous across all three domains of life and seemed at one point to be the only such ATP-dependent protease. Analysis of the T. maritima genome revealed a homolog to the Escherichia coli Lon, TM1633, referred to here as LonA. However, a second open reading frame, TM1869, was also annotated as a possible Lon homolog, designated LonB, which was previously not identified. LonA has both the active site and ATP-binding site motifs typically noted in other versions of Lon. LonB contains the active site region (with the conserved serine residue), but not the ATP-binding site (Figure 1). Homologs to both LonA and LonB were noted in several bacterial genomes. However, euryarchaeal genomes appear to encode only a single LonB homolog. In Pyrococcus spp., the lon gene contains an intein (http://www.neb.com/neb/frame_tech.html), a self-splicing element functional at the protein level. Whether this impacts its regulation or the expression of the encoded protein is unknown. No Lon homolog could be identified in the S. solfataricus genome. The archaeal Lons also have one or two putative transmembrane regions, which are absent in the Eubacteria, suggesting that they may be membrane-associated. LonA and LonB are distinct enzymes, as is readily apparent when a phylogenetic tree is constructed based on their amino acid sequences (Figure 2). Although it is clear that LonA and LonB are distinct enzymes, their metabolic roles and biochemical specifics are unknown.
Figure 1.
Multiple sequence alignment using CLUSTALX of the ATP-binding domain and active site region of the LonA and LonB homologs. Abbreviations: Ecoli = Escherichia coli; Paer = Pseudomonas aeruginosa; Msmeg = Mycobacterium smegmatis; Bsub = Bacillus subtilis; Ttherm = Thermus thermophilus; Aaeol = Aquifex aeolicus; Tmar = Thermotoga maritima; Pfur = Pyrococcus furiosus; and Tpacid = Thermoplasma acidophilum.
Figure 2.
Phylogenetic tree of Lon proteins.
The FtsH (or HflB) proteinase
FtsH (or HflB) is an ATP-dependent proteinase with a zinc metalloprotease motif that is present in Bacteria, mitochondria, and chloroplasts, but not in Archaea (Schumann 1999, Langer 2000). Unlike the proteasome, Lon protease and Clp protease, which are cytoplasmic, the FtsH protein is anchored to the cytoplasmic membrane through two transmembrane regions. Among hyperthermophiles, FtsH has been located only in the bacteria Aquifex aeolicus and T. maritima (see Table 1). Although Archaea lack FtsH, we speculate that the archaeal Lon, which has one to two putative transmembrane regions, may replace FtsH from a metabolic standpoint.
The Clp family of proteases
The Clp family occurs in both the eubacteria and eukaryotes, including hyperthermophilic bacteria, but appears to be absent in hyperthermophilic archaea. Two types of Clp proteases are known, with either ClpP or ClpQ (HslV) as the proteolytic subunit. For full proteolytic activity, ClpP and ClpQ must associate with their respective ATPase subunits. In the case of ClpP this can be either ClpX or ClpA ATPases, whereas ClpQ associates with ClpY (HslU) ATPase (Porankiewicz et al. 1999). Both the ClpP and ClpQ proteases, as well as their respective ATPase subunits ClpC (three homologs), ClpX and ClpY were identified in the genomes of T. maritima and A. aeolicus (Deckert et al. 1998). In both T. maritima and A. aeolicus, the clpP gene is preceded by a putative trigger factor. The trigger factor may be involved in the export of proteins, acting as a chaperone to keep them in an open conformation. Moreover, ClpP in the A. aeolicus genome is followed by ClpX. In T. maritima, the Clp ATPases are scattered throughout the genome and are not linked to other proteases. However, ClpC-1 in T. maritima (TM0198) is linked to radA, which is involved in DNA repair. We postulate that these genes are involved in stress response and are coordinately regulated. The genes encoding ClpQ and ClpY are linked on the T. maritima genome and are likely co-transcribed, whereas they occur separately in the A. aeolicus genome. As more is learned about gene regulation in these two hyperthermophiles, the significance of these alternative arrangements for ClpQ and ClpY should become clearer.
In E. coli,expression of lon, clpP and the ATPase subunits clpX and clpB, are all σ32-dependent (Bukau 1993), whereas in gram-positive Bacillus subtilis and Lactococcus lactis, the heat-shock response involves at least three classes of heat-inducible genes (Hecker et al. 1996). It has recently been found that, in gram-positive bacteria, clpP is under the control of a novel regulator, CtsR, belonging to Class III, which is σB-independent and lacks the cis-acting CIRCE operator sequence (Hecker et al. 1996). Analysis of the T. maritima genome reveals 70 putative transcriptional regulators that have moderate degrees of identity to regulators from both gram-positive and gram-negative sources (Nelson et al. 1999). Among these are homologs to the E. coli sigma E (rpoE) and the heat-shock operon repressor (HcrA) from B. subtilis (Nelson et al. 1999). However, homologs to E. coli σ32 and the gram-positive regulator CtsR could not be identified. It is possible that T. maritima uses a novel mechanism, or a regulatory pathway that is a hybrid of that in gram-positive and gram-negative organisms.
The proteasome
The 20S proteasome, or multicatalytic proteinase (MCP), is a cylindrically shaped protease found in the Archaea, Eukarya and the gram-positive actinomycetes (Bochtler et al. 1999, Barber and Ferry 2001). The lack of the proteasome in other bacteria suggests that actinomycetes acquired the protease through lateral gene transfer (Lupas et al. 1997). Although most bacteria lack a version of the proteasome, they contain the related complex ClpQY (or HslVU), which shares a similar fold and catalytic mechanism with the proteasome (Bochtler et al. 1997). The proteasome from the thermophilic archaeon Thermoplasma acidophilum yielded the first structure of the proteasome and it has since become a prototype for the three-dimensional structure and topology of the molecule (Lowe et al. 1995). Native versions of the archaeal proteasome have been isolated and characterized from Methanosarcina thermophila, Methanococcus jannaschii and P. furiosus and appear to share many structural and biochemical properties with the T. acidophilum proteasome (Maupin-Furlow and Ferry 1995, Bauer et al. 1996, Wilson et al. 2000). The 20S structure comprises four heptametrical rings stacked on top of one another (Rechsteiner et al. 1993). Each ring comprises either α- or β-type subunits, arranged in the order α7β7β7α7, with a centralized hollow channel running the entire length of the complex (DeMartino and Slaughter 1999). To safeguard against unwanted protein degradation, the proteasome confines proteolytic activity to the interior region of this self-compartmentalized structure (DeMartino and Slaughter 1999, Goldberg 2000). Although the archaeal 20S proteasome functions as a discrete protease in vitro, it is not known if the 26S proteasome is the only functional form in vivo (Maupin-Furlow et al. 2000). Archaeal proteasomes contain various peptidase activities; most have only chymotrypsin-like activity, although some also have high trypsin-like or caspase-like activity. The archaeal proteasome acts in a processive manner, chopping protein substrates at multiple places to yield peptide fragments of three to 30 amino acids in length (Kisselev et al. 1998). Although the physiological role of the archaeal proteasome is unclear, inhibitor-based studies show that T. acidophilum cells can proliferate without a functional proteasome under normal growth conditions, but cannot grow without proteasome activity under heat shock conditions (Ruepp et al. 1998).
All archaeal genomes sequenced to date contain homologs of the 20S proteasome core structure, including members of both Crenarchaeota and Euryarchaeota (see Tables 1 and 2). The genomes of Aeropyrum pernix, Pyrococcus spp. and S. solfataricus each contain two different β subunit homologs (see Tables 1 and 2), and two different α subunits have been identified in the halophilic archaeon Haloferax volcanii (Wilson et al. 1999). In Archaea, the genes encoding the α and β proteasome subunits appear to be transcribed as a part of independent operons that have conserved gene organization, whereas the genes surrounding pan (proteosome activating nucleotidase) do not appear to be conserved (Maupin-Furlow et al. 2000). In addition, the archaeal proteasome appears to be a part of a superoperon containing subunits of the exosome, indicating a possible functional link between RNA processing and proteolysis in this domain (Koonin et al. 2001).
It is clear that hyperthermophiles, like their mesophilic counterparts, have a full complement of ATP-dependent proteases at their disposal. However, the proteases differ between the two domains, the proteasome and Lon proteases being present in Archaea and the Lon, Clp and FtsH being present in Eubacteria. Although much is known about ATP-dependent proteases and their metabolic roles in mesophilic eubacteria, one cannot assume that they have identical roles or regulation patterns in hyperthermophilic bacteria. Moreover, little is known about the metabolic roles and regulation of the proteasome and Lon in Archaea.
ATP-independent proteases in hyperthermophiles
Most heterotrophic hyperthermophiles can grow on proteinaceous substrates as primary carbon and energy sources. Such substrates must initially be acted on by extracellular proteases, which may or may not be cell-associated. The products of extracellular hydrolysis are transported into the cell, presumably by an ABC-type transporter, where they are further broken down to individual amino acids by the concerted action of intracellular proteases and peptidases. It is thought that peptides can be oxidized to CO2 in Thermoproteus tenax, Archaeoglobus fulgidus and S. solfataricus, with sulfur, thiosulfate, sulfate, oxygen, nitrite and nitrate serving as terminal electron acceptors (Schönheit and Schäfer 1995). In species from the genera Thermococcus, Pyrococcus, Thermotoga, Desulfurococcus and Pyrodictum, peptides are likely fermented to free acids, such as acetate, isovalerate, butyrate and phenylpyruvate, generating ATP by substrate-level phosphorylation (Schäfer et al. 1993, Schönheit and Schäfer 1995). Parts of these pathways can be constructed by analysis of the genomes of these organisms, although the details of metabolic schemes involved in peptide fermentation in these organisms are unknown.
Proteases and peptidases presumably involved in the initial steps in protein and peptide utilization have been isolated from hyperthermophilic archaea, including Pyrococcus spp. (Halio et al. 1996, Voorhorst et al. 1996, Chang et al. 2001), Thermococcus stetteri (Klingeberg et al. 1995), A. pernix (Sako et al. 1997) and P. abyssi (Dib et al. 1998) (see Table 3). The only protease isolated from T. maritima thus far is a homomultimeric protease that has moderate amino acid sequence identity to bacteriocins from mesophilic bacteria (Hicks et al. 1998). Within the classical classification scheme for proteases, namely, serine, aspartic, metallo and cysteine, the majority of the enzymes characterized to date from hyperthermophiles has been extracellular, and belongs to the serine class. Although these enzymes have been well characterized biochemically, little is known about their metabolic significance and even less about their regulation.
Table 3.
Proteases and peptidases isolated from hyperthermophilic microorganisms. Abbeviations: (NP) = recombinant form, not purified from host; M = metalloprotease; Ser = serine; Cys = cysteine; E = extracellular; I = intracellular; and MA = membrane associated.
| Organism | Protease | Type | Topt (°C) | pHopt | MW (kDa) | Location | Reference |
| Crenarchaeota | |||||||
| Sulfolobus acidocaldarius | Thermopsin | Acid | 90 | 2 | Calc. 32.6, App 46–51 | E | Fusek et al. 1990, Lin et al. 1991 |
| Sulfolobus solfataricus | Aminopeptidase | M (Co2+/Mn2+) | 75 | 6.75 | 4 × 80 (320) | I | Hanner et al. 1990 |
| S. solfataricus | Carboxypeptidase | M (Zn2+) | 85 | 5.5–7/9 | 4 × 42 (170) | I | Colombo et al. 1992, 1995 |
| S. solfataricus | Proteinase I | Ser | > 90 | 6.5–8 | 2 × 54 (118) | I | Burlini et al. 1992 |
| Aeropyrum pernix K1 | Aeropyrolysin | M | 100 (– CaCl2) | 6.0–8.0 | 52 | E | Sako et al. 1997 |
| 110 (+ CaCl2) | |||||||
| A. pernix K1 | Pernilase | Ser | 90 | 9 | 50 | I | Chavez Croocker et al. 1999 |
| Desulfurococcus mucosus | Archaelysin | Ser | 90–95 | 7.2 | 43–54 | E | Cowan et al. 1987 |
| Pyrobaculum aerophilum | Aerolysin (NP) | Ser | E? | Volkl et al. 1994 | |||
| Euryarchaeota | |||||||
| Pyrococcus abyssi st 549 | Protease from Pab (PPA) | Ser | 95 | 9 | 60, 105, 150 | E | Dib et al. 1998 |
| Pyrococcus furiosus | Carboxypeptidase (PfuCP) | M (Co2+) | 90–100 | 6.2–6.5 | 2 × 58–9 (128) | I | Cheng et al. 1999 |
| P. furiosus | Deblocking aminopeptidase (DAP) | 10–12 subunits | I | Tsunasawa 1998 | |||
| (size not reported) | |||||||
| P. furiosus | Methionine aminopeptidase | M (Co2+) | ~90 | 7.0–8.0 | 32.74 | I | Tsunasawa et al. 1997, Tahirov et al. 1998 |
| (PfMAP) (NP) | |||||||
| P. furiosus | Proline dipeptidase/prolidase | M (Co2+) | 100 | 7 | 2 × 39.4–42 (79) | I | Ghosh et al. 1998 |
| P. furiosus | Prolyl endopeptidase/oligopeptidase | Ser | 85–90 | 7.0–8.0 | Calc. 70–1, App 59 | I | Robinson et al. 1995, Harwood et al. 1997, |
| (POP) | Harris et al. 2001 | ||||||
| P. furiosus | Protease I (PfpI) | Cys | 105 | 7 | 18.8 subunit, 60 | I | Blumentals et al. 1990, Halio et al. 1996, 1997, |
| and 120 native | Du et al. 2000, Chang et al. 2001 | ||||||
| P. furiosus | Pyrolysin | Ser | 115 | 6.5–10.5 | 150 | MA | Voorhorst et al. 1996 |
| P. furiosus | Pyrrolidone carboxyl peptidase | Cys | ~90 | 6.0–9.0 | 4 × 22.8 (96.3) | I | Tsunasawa et al. 1998 |
| (PfPCP) (NP) | |||||||
| Pyrococcus horikoshii | Aminopeptidase (NP) | 95 | 7.5 | X × 36.9 (440) | I | Ando et al. 1999 | |
| Thermococcus kodakaraensis | Subtilisin-like protease (NP) | Ser | 80 | 9.5 | 43.8 | E? | Kannan et al. 2001 |
| T. kodakaraensis | Thiol protease (TT) | Cys | 110 | 7 | 44 | E | Morikawa et al. 1994 |
| Thermococcus litoralis | Pyrrolidone carboxyl peptidase (NP) | Cys | 70 or 75 | 4 × 24 (96) | I | Singleton et al. 1999, 2000 | |
| Thermococcus stetteri | Stetterlysin | Ser | MA | Voorhorst et al. 1997 | |||
| T. stetteri | SDS-resistant protease | Ser | 85 | 8.5–9 | 68 | E | Klingeberg et al. 1995 |
| Staphylothermus marinus | STABLE protease | Ser | 90 | 9 | 150 | MA | Peters et al. 1995, Mayr et al. 1996 |
| Thermoplasma acidophilum | Tricorn protease (TRI) | Ser | 65 | 8.5–8.8 | 120 (monomer); | I | Tamura et al. 1996 |
| 720 (hexamer); | |||||||
| 14,600 (icosahedral | |||||||
| capsid/20 hexamers) | |||||||
| Bacteria | |||||||
| Aquifex pyrophilus | Serine-type protease (NP) | Ser | 85 | 7.0–9.0 | 43 | MA | Choi et al. 1999 |
| Fervidobacterium pennivorans | Fervidolysin | Ser | 80 | 10 | 73 (precursor); | MA | Friedrich and Antranikian 1996, Kluskens |
| 58 (mature protease); | et al. 2001 | ||||||
| 130 (Friedrich – | |||||||
| native purif.) | |||||||
| Thermotoga maritima | Maritimacin | 90–93 | 6.0–9.0 | X × 31 (> 669) | I | Hicks et al. 1998, 2001 | |
Proteolysis in hyperthermophilic bacteria is poorly understood, despite the availability of complete genomic sequences for T. maritima (Nelson et al. 1999) and A. aeolicus (Deckert et al. 1998). To date, only two proteases (Hicks et al. 1998, Choi et al. 1999) and a leucine aminopeptidase (Khan et al. 2000) have been characterized biochemically from hyperthermophilic bacteria. A 43-kDa serine protease was identified in Aquifex pyrophilus using a sequence tag specific for serine proteases (Choi et al. 1999). The gene encoding the protease was sequenced and found to contain a putative signal sequence. The protease was identified in the cell wall fraction of A. pyrophilus, demonstrating that the protease is expressed in the native organism and that it is exported from the cell. However, the role of the protease is unclear because Aquifex spp. appear unable to use peptides as a carbon or nitrogen source (Deckert et al. 1998). Choi et al. (1999) suggested that the protease is associated with the cellular S-layer, but whether it is involved in protein degradation or is involved in S-layer formation is unclear. Similarly, in T. maritima, analysis of the genome identified about 35 proteases and peptidases, 12 of which may be exported (see Tables 1 and 2). However, growth of T. maritima on peptides as sole carbon and energy sources has not been reported.
Among the hyperthermophiles, proteolysis has been best studied in members of the order Thermococcales. These organisms can use peptides as sole carbon and energy sources and this is reflected in a range of proteolytic activities in their cell extracts and confirmed by genomic sequence analysis. Initial efforts to study P. furiosus indicated that the organism was highly proteolytic (Blumentals et al. 1990, Eggen et al. 1990, Connaris et al. 1991). Analysis of the P. furiosus genome has thus far revealed the presence of about 40 genes encoding proteases, protease subunits, or peptidases (Table 1). Nine of the identified proteases contain a putative signal sequence, suggesting that they are exported from the cell. A comparative genomics approach was also used to compare P. furiosus, P. abyssi and P. horikoshii in an effort to access the proteolytic inventory within this narrow phylogenetic range of the Thermococcales (Table 1). Similar to P. furiosus, both P. abyssi and P. horikoshii are highly proteolytic, with 34 genes encoding proteolytic enzymes detected in their genome sequences. It is apparent that, despite their close phylogenetic relationship, there are distinct differences between these organisms in their respective proteolytic inventories. Pyrococcus furiosus has four proteases (pf_699579, pf_1553191, pf_1757236 and pf_1136394) and three aminopeptidases (pf_1898123, pf_1902688 and pf_1906416) that are absent from P. abyssi and P. horikoshii. There is only one protease unique to P. abyssi (CAAX Prenyl protease, PAB0555), whereas no proteases yet identified are unique to P. horikoshii. Moreover, there is a clear distinction between the pyrolysin proteases in the Thermococcales. The pyrolysin from P. furiosus is only 17% identical to the pyrolysin-like proteases of P. abyssi and P. horikoshii, and 33% identical to stetterolysin from Thermococcus stetteri, whereas the pyrolysin-like proteases of P. abyssi and P. horikoshii are 69% identical. Differences in proteolytic content of Thermococcales can also be seen by zymogram analysis. For example, Figure 3 shows that the extracellular proteases expressed by P. furiosus, P. abyssi, Thermococcus profundus, Thermococcus peptonphilius and Thermotoga maritima vary when grown in the same medium at their respective temperature optima with tryptone (5 g l–1) as the primary carbon and energy source.
Figure 3.
Extracellular proteolytic inventory from various hyperthermophiles. Pyrococcus furiosus and P. abyssi were grown at 95 °C, and T. profundus, T. peptonophilis and Thermotoga maritima were grown at 80 °C. The cells were removed after 14 h of growth and the extracellular enzymes were precipitated by addition of ammonium sulfate to 80%. Two µg of total extracellular protein was loaded onto a 12% SDS-polyacrylamide gel containing gelatin.
Several extracellular (Morikawa et al. 1994, Klingeberg et al. 1995, Dib et al. 1998, Kannan et al. 2001) and membrane-associated proteases (Voorhorst et al. 1996, 1997) have been characterized biochemically from both Thermococcus and Pyrococcus spp. With the exception of the thiol protease from T. kodakaraensis, all the extracellular and membrane-associated proteases have been classified as serine proteases. The proteases range in size from 40 to 68 kDa and are monomeric, with the exception of the P. abyssi protease, which is multimeric. The serine protease from T. kodakaraensis has broad substrate specificity, cleaving at the carboxyl termini of Tyr, Phe, Leu, Gln, His, Thr, Ser and Ala (Kannan et al. 2001). The other serine proteases from Thermococcus stetteri and P. abyssi have fairly specific substrate specificity, preferring Arg/Phe and Aromatic/Leu at the P1 position, respectively (Klingeberg et al. 1995, Dib et al. 1998). Substrate specificity of the thiol protease was not determined.
An intracellular protease identified in P. furiosus (Blumentals et al. 1990) and designated Pyrococcus furiosus protease I (PfpI) was found to be based on a single 18.8-kDa subunit (Halio et al. 1997). The gene encoding this subunit has putative homologs in other cells and microorganisms from the three domains of life, including M. jannaschii, B. subtilis, E. coli and Homo sapiens, but not T. maritima or Saccharomyces cerevisiae. In vitro, PfpI occurs in at least three functional forms, a trimer, a hexamer and a dodecamer, and is most active as a dodecamer (Chang et al. 2001). The physiological role of PfpI is unclear, but it complements the proteasome in P. furiosus; in vitro, a synergistic relationship between the two proteases has been noted (Chang et al. 2001). Because P. furiosus lacks the tricorn protease (Table 1), we speculate that PfpI assumes this role in P. furiosus. It also appears to be a predominant protease in P. furiosus based on zymogram analyses. The three-dimensional structure of the PfpI homolog in P. horikoshii (PhpI) (90% identical at the amino acid level) was recently reported (Du et al. 2000). The structure was consistent with previously reported biophysical information: PhpI is a dodecamer consisting of two identical six-member rings, each with axes of symmetry such that it consists of a dimer of trimers or a trimer of dimers. This supports the observation that, in vitro, it exists in at least three functional forms (P.M. Hicks, North Carolina State Univ., and R.M. Kelly, unpublished data). Furthermore, even though PfpI/PhpI are not ATP-dependent, the structure of PhpI suggests a similar barrel-like compartmentalization of the active site, reminiscent of the 20S proteasome and ClpP. The possible relationship of PfpI/PhpI and other ATP-independent proteases with protected active sites to ATP-dependent proteases needs to be examined, especially with respect to the evolutionary significance of the energetic requirement.
Sulfolobus solfataricus can use proteinaceous compounds as primary carbon and energy sources, and analysis of the S. solfataricus genome revealed the presence of 37 genes encoding proteases, protease subunits, or peptidases (Table 2). The tricorn protease as well as the interacting F factors F1, F2 and F3, which are aminopeptidases, were also identified (She et al. 2001). Although it has not been characterized in S. solfataricus, the tricorn protease has been well characterized in the thermophilic archaeon Thermoplasma (Tamura et al. 1998). Tricorn protease, in conjunction with its three interacting factors, degrades oligopeptides in a sequential manner, yielding free amino acids.
Two proteases have been characterized in Sulfolobus spp.; a heat-stable intracellular serine protease from S. solfataricus (Burlini et al. 1992), and the acid protease thermopsin from S. acidocaldarius (Fusek et al. 1990, Lin et al. 1991). The protease from S. solfataricus has a subunit molecular mass of 54 kDa and the active form is 118 kDa, suggesting that it exists as a dimer. The enzyme has chymotrypsin-like specificity, preferring aromatic or bulky aliphatic amino acids, but differs from chymotrypsin in being unable to digest natural substrate proteins like insulin chains A and B (Burlini et al. 1992). Thermopsin has a predicted molecular mass of 32.6 kDa, but was found to be a monomer with a molecular mass of 46 kDa. The difference in molecular mass is likely because thermopsin is a glycoprotein (Lin et al. 1991). Thermopsin is capable of degrading various protein substrates, such as hemoglobin, ovalbumin, bovine serum albumin and insulin chain B (Fusek et al. 1990). The enzyme has a rather broad substrate specificity but, similar to pepsin, prefers hydrophobic residues to flank the cleavage site.
Proteases specific to hyperthermophiles
Genome sequence analysis shows that there is considerable conservation of certain proteases across all domains of life and growth temperature ranges. However, some proteases appear unique or more common to hyperthermophiles. For example, three clostripain-related proteases appear to be found only in T. maritima. These proteases have limited homology (32% identical in a 125 amino acid region) to the heterodimeric cysteine endoprotease from Clostridium histolyticum (Dargatz et al. 1993). However, given the difficulties in cloning and expressing active forms of hyperthermophilic proteases in mesophilic hosts, biophysical and biochemical information for these enzymes may need to rely to a great extent on direct purification.
There are examples of proteases that, from a structural or functional perspective, may be unique to hyperthermophiles. For example, the homomultimeric protease in T. maritima based on an approximately 31-kDa subunit has significant homology at the amino acid sequence level to a gene encoding a mesophilic bacteriocin, linocin M18; this protease has been tentatively named maritimacin (Hicks et al. 1998). A putative homolog to this protease has also been identified in the genome sequence of P. furiosus, but the enzyme appears to be absent from the rest of the Archaea (Table 1). Linocin M18, a multimeric assembly of a single 31-kDa subunit isolated from Bevibacterium linens, was found to be antagonistic to a wide spectrum of mesophilic coryneform and other gram-positive bacteria (Valdes-Stauber and Scherer 1996). However, the physiological function of this protein in these two hyperthermophilic organisms and its relationship to bacteriocins are unknown. Initial efforts to screen the purified T. maritima protease for bacteriocin-like activity focused on several hyperthermophiles that are readily cultured, including hyperthermophilic archaea. However, no significant antagonistic activity was found, although the presence of the putative protease seemed to extend the lag phase for Thermococcus litoralis (Hicks et al. 2001).
Pyrolysin is a cell envelope-associated protease that has N-terminal sequence homology with subtilisin-like serine proteases purified from P. furiosus (Voorhorst et al. 1996). The enzyme exhibits endopeptidase activity and may be involved in the first step of protein utilization during proteolytic growth (Voorhorst et al. 1996). Pyrolysin exists as a mosaic of domains shared by other proteases containing 1398 amino acid residues, including a conserved pre-proenzyme region, a catalytic domain of 500 residues, and a large C-terminal extension, making it one of the largest known serine proteases (de Vos et al. 2001). Pyrolysin exhibits highest identity with the catalytic domain of the eukaryal tripeptidyl peptidases II, a subgroup of the subtilisin-like proteases (Voorhorst et al. 1996). Although the catalytic domain of pyrolysin is found in a putative subtilase from P. furiosus, the intact gene is absent from the genome sequences of the rest of the Archaea, including P. horikoshii and P. abyssi. However, a gene was identified in the genome sequences of P. horikoshii (PH0310) and P. abyssi (PAB1252), which includes the conserved pre-pro- and C-terminal sequences of pyrolysin, but contains a putative protease with a thiol-protease catalytic domain rather than serine protease activity (de Vos et al. 2001).
Another surface layer bound subtilisin-like protease, STABLE, has been identified in Staphylothermus marinus (Mayr et al. 1996) that has some relationship to pyrolysin in that specific motifs are common to both proteases. This protease presumably digests and provides peptides to this organism, which grows by sulfur-dependent, peptide fermentation.
Evolutionary aspects of proteolytic processes
Given that hyperthermophiles arguably represent an early evolutionary linkage, the question concerning the minimum set of proteases required for cellular function in these organisms arises. About half of the genes contained in hyperthermophilic genomes are still unassigned in terms of function (Nelson et al. 1999), and it seems certain that as yet undetected proteases are among those unidentified genes. About 60–70 putative and actual proteases/peptidases can be gleaned from the E. coli genome sequence by informatics techniques similar to those used here for the hyperthermophiles (K.R. Shockley and R.M. Kelly, unpublished data); this is about twice as many as noted in each of the five hyperthermophiles examined here (Table 2). We also note that the E. coli genome is about 2.5-fold larger than the pyrococcal and T. maritima genomes, which may relate to differences in protease inventory. However, S. solfataricus and E. coli have comparably sized genomes but differ significantly in the numbers of putative and confirmed proteases. Information about the expression and activation of specific proteases under various environmental conditions is needed to resolve the relationship between genotype and phenotype for each organism. The biochemical properties of specific proteases in these organisms will have to be reconciled with their regulation to provide some perspective on the global regulation of proteolysis.
It is intriguing to consider the origin and development of proteases based on the multimeric assembly of small (~20–30 kDa) subunits into complex ring-like structures. This is the basis for the proteasome and Clp proteases, both of which are ATP-dependent. Such structures help sequester the active sites in these proteases, presumably to avoid unwanted protein turnover. The PfpI from P. furiosus and its homologs are based on a similar structural organization (~19-kDa subunits arranged into two six member rings) yet have no ATP-dependence (Du et al. 2000). Furthermore, there may be an additional structural relationship between these proteases, given that antibodies raised against the eukaryotic proteasome and E. coli ClpP recognized PfpI in Western blots (Halio et al. 1996). The evolutionary significance of these multi-subunit proteases and the complexity introduced through ATP-dependence merits further examination. It will be interesting to determine whether various proteolytic phenotypes recruit specific sets of proteases to certain tasks involving protein turnover under normal and stressed conditions and how the interplay between ATP-dependence and ATP-independence is regulated. If it turns out that hyperthermophiles contain fewer proteases than other cells and organisms, they may provide an interesting perspective on the complex nature of protease function and regulation.
Acknowledgments
This work was supported in part by grants from the National Science Foundation, the U.S. Department of Energy, Energy Biosciences Program and Syngenta Corporation (Research Triangle Park, NC).
References
- R1.Allison C., Macfarlane G.T. Regulation of protease production in Clostridium sporogenes . Appl. Environ. Microbiol. 1990;56:3485–3490. doi: 10.1128/aem.56.11.3485-3490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Arsene F., Tomoyasu T., Bukau B. The heat shock response of Escherichia coli . Int. J. Food Microbiol. 2000;55:3–9. doi: 10.1016/s0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- R3.Barber R.D., Ferry J.G. Archaeal proteasomes. Methods Enzymol. 2001;330:413–424. doi: 10.1016/s0076-6879(01)30393-2. [DOI] [PubMed] [Google Scholar]
- R4.Bauer M.W., Halio S.B., Kelly R.M. Proteases and glycosyl hydrolases from hyperthermophilic microorganisms. Adv. Protein Chem. 1996;48:271–310. doi: 10.1016/s0065-3233(08)60364-2. [DOI] [PubMed] [Google Scholar]
- R5.Blumentals I.I., Robinson A.S., Kelly R.M. Characterization of sodium dodecyl sulfate-resistant proteolytic activity in the hyperthermophilic archaebacterium Pyrococcus furiosus . Appl. Environ. Microbiol. 1990;56:1992–1998. doi: 10.1128/aem.56.7.1992-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R6.Bochtler M., Ditzel L., Groll M., Huber R. Crystal structure of heat shock locus V (HslV) from Escherichia coli . Proc. Natl. Acad. Sci. USA. 1997;94:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Bochtler M., Ditzel L., Groll M., Hartmann C., Huber R. The proteasome. Annu. Rev. Biophys. Biomol. Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- R8.Brock T.D., Brock K.M., Belly R.T., Weiss R.L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- R9.Bukau B. Regulation of the Escherichia coli heat-shock response. Mol. Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- R10.Burlini N., Magnani P., Villa A., Macchi F., Tortora P., Guerritore A. A heat-stable serine proteinase from the extreme thermophilic archaebacterium Sulfolobus solfataricus . Biochim. Biophys. Acta. 1992;1122:283–292. doi: 10.1016/0167-4838(92)90406-4. [DOI] [PubMed] [Google Scholar]
- R11.Chang L.S., Hicks P.M., Kelly R.M. Protease I from Pyrococcus furiosus . Methods Enzymol. 2001;330:403–413. doi: 10.1016/s0076-6879(01)30392-0. [DOI] [PubMed] [Google Scholar]
- R12.Choi I.G., Bang W.G., Kim S.H., Yu Y.G. Extremely thermostable serine-type protease from Aquifex pyrophilus. Molecular cloning, expression, and characterization. J. Biol. Chem. 1999;274:881–888. doi: 10.1074/jbc.274.2.881. [DOI] [PubMed] [Google Scholar]
- R13.Connaris H., Cowan D., Sharp R. Heterogeneity of proteinases from the hyperthermophilic archeobacterium Pyrococcus furiosus . J. Gen. Microbiol. 1991;137:1193–1199. [Google Scholar]
- R14.Dargatz H., Diefenthal T., Witte V., Reipen G., von Wettstein D. The heterodimeric protease clostripain from Clostridium histolyticum is encoded by a single gene. Mol. Gen. Genet. 1993;240:140–145. doi: 10.1007/BF00276893. [DOI] [PubMed] [Google Scholar]
- R15.de Vos W.M., Voorhorst W.G., Dijkgraaf M., Kluskens L.D., van der Oost J., Siezen R.J. Purification, characterization, and molecular modeling of pyrolysin and other extracellular thermostable serine proteases from hyperthermophilic microorganisms. Methods Enzymol. 2001;330:383–393. doi: 10.1016/s0076-6879(01)30390-7. [DOI] [PubMed] [Google Scholar]
- R16.Deckert G., Warren P.V., Gaasterland T., et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus . Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- R17.DeMartino G.N., Slaughter C.A. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- R18.Dib R., Chobert J.M., Dalgalarrondo M., Barbier G., Haertle T. Purification, molecular properties and specificity of a thermoactive and thermostable proteinase from Pyrococcus abyssi, strain st 549, hyperthermophilic archaea from deep-sea hydrothermal ecosystem. FEBS Lett. 1998;431:279–284. doi: 10.1016/s0014-5793(98)00782-0. [DOI] [PubMed] [Google Scholar]
- R19.Du X.L., Choi I.G., Kim R., Wang W.R., Jancarik J., Yokota H., Kim S.H. Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-Å resolution. Proc. Natl. Acad. Sci. USA. 2000;97:14079–14084. doi: 10.1073/pnas.260503597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Eggen R.I., Geerling A.C., Watts J., de Vos W.M. Characterization of pyrolysin, a hyperthermoactive serine protease from the archaebacterium Pyrococcus furiosus . FEMS Microbiol. Lett. 1990;71:17–20. [Google Scholar]
- R21.Erauso G., Reysenbach A.-L., Godfroy A., et al. Pyrococcus abyssi sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Arch. Microbiol. 1993;160:338–349. [Google Scholar]
- R22.Fiala G., Stetter K.O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 °C. Arch. Microbiol. 1986;161:168–175. [Google Scholar]
- R23.Fusek M., Lin X.L., Tang J. Enzymic properties of thermopsin. J. Biol. Chem. 1990;265:1496–1501. [PubMed] [Google Scholar]
- R24.Goldberg A.L. Probing the proteasome pathway. Nat. Biotechnol. 2000;18:494–496. doi: 10.1038/75349. [DOI] [PubMed] [Google Scholar]
- R25.Gonzalez J.M., Masuchi Y., Robb F.T., Ammerman J.W., Yanagibayashi M., Tamaoka J., Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998;2:123–130. doi: 10.1007/s007920050051. [DOI] [PubMed] [Google Scholar]
- R26.Gottesman S. Proteases and their targets in Escherichia coli . Annu. Rev. Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- R27.Gottesman S. Regulation by proteolysis: developmental switches. Curr. Opin. Microbiol. 1999;2:142–147. doi: 10.1016/S1369-5274(99)80025-3. [DOI] [PubMed] [Google Scholar]
- R28.Guedon E., Renault P., Ehrlich S.D., Delorme C. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 2001;183:3614–3622. doi: 10.1128/JB.183.12.3614-3622.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Halio S.B., Blumentals I.I., Short S.A., Merrill B.M., Kelly R.M. Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus . J. Bacteriol. 1996;178:2605–2612. doi: 10.1128/jb.178.9.2605-2612.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R30.Halio S.B., Bauer M.W., Mukund S., Adams M.W.W., Kelly R.M. Purification and characterization of intracellular protease PfpI from the hyperthermophilic archaeon Pyrococcus furiosus . Appl. Environ. Microbiol. 1997;63:289–295. doi: 10.1128/aem.63.1.289-295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R31.Hecker M., Schumann W., Volker U. Heat-shock and general stress response in Bacillus subtilis . Mol. Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- R32.Hicks P.M., Rinker K.D., Baker J.R., Kelly R.M. Homomultimeric protease in the hyperthermophilic bacterium Thermotoga maritima has structural and amino acid sequence homology to bacteriocins in mesophilic bacteria. FEBS Lett. 1998;440:393–398. doi: 10.1016/s0014-5793(98)01451-3. [DOI] [PubMed] [Google Scholar]
- R33.Hicks P.M., Chang L.S., Kelly R.M. Homomultimeric protease and putative bacteriocin homolog from Thermotoga maritima . Methods Enzymol. 2001;330:455–460. doi: 10.1016/s0076-6879(01)30397-x. [DOI] [PubMed] [Google Scholar]
- R34.Huber R., Langworthy T.A., König H., Thomm M., Woese C.R., Sleytr U.B., Stetter K.O. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 °C. Arch. Microbiol. 1986;144:324–333. [Google Scholar]
- R35.Kannan Y., Koga Y., Inoue Y., Haruki M., Takagi M., Imanaka T., Morikawa M., Kanaya S. Active subtilisin-like protease from a hyperthermophilic archaeon in a form with a putative prosequence. Appl. Environ. Microbiol. 2001;67:2445–2452. doi: 10.1128/AEM.67.6.2445-2452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R36.Kawarabayasi Y., Sawada M., Horikawa H., et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- R37.Khan A.R., Nirasawa S., Kaneko S., Shimonishi T., Hayashi K. Characterization of a solvent resistant and thermostable aminopeptidase from the hyperthermophillic bacterium, Aquifex aeolicus . Enzyme Microb. Technol. 2000;27:83–88. doi: 10.1016/s0141-0229(00)00194-0. [DOI] [PubMed] [Google Scholar]
- R38.Kisselev A.F., Akopian T.N., Goldberg A.L. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J. Biol. Chem. 1998;273:1982–1989. doi: 10.1074/jbc.273.4.1982. [DOI] [PubMed] [Google Scholar]
- R39.Klingeberg M., Galunsky B., Sjoholm C., Kasche V., Antranikian G. Purification and properties of a highly thermostable, sodium dodecyl sulfate-resistant and stereospecific proteinase from the extremely thermophilic archaeon Thermococcus stetteri . Appl. Environ. Microbiol. 1995;61:3098–3104. doi: 10.1128/aem.61.8.3098-3104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R40.Koonin E.V., Wolf Y.I., Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R41.Langer T. AAA proteases: cellular machines for degrading membrane proteins. TIBS. 2000;25:247–251. doi: 10.1016/s0968-0004(99)01541-8. [DOI] [PubMed] [Google Scholar]
- R42.Lin X., Fusek M., Tang J. Thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius . Adv. Exp. Med. Biol. 1991;306:255–257. doi: 10.1007/978-1-4684-6012-4_29. [DOI] [PubMed] [Google Scholar]
- R43.Lowe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- R44.Lupas A., Flanagan J.M., Tamura T., Baumeister W. Self-compartmentalizing proteases. Trends Biochem. Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- R45.Maupin-Furlow J.A., Ferry J.G. A proteasome from the methanogenic archaeon Methanosarcina thermophila . J. Biol. Chem. 1995;270:28617–28622. doi: 10.1074/jbc.270.48.28617. [DOI] [PubMed] [Google Scholar]
- R46.Maupin-Furlow J.A., Wilson H.L., Kaczowka S.J., Ou M.S. Proteasomes in the Archaea: from structure to function. Front. Biosci. 2000;5:837–865. doi: 10.2741/furlow. [DOI] [PubMed] [Google Scholar]
- R47.Maurizi M.R. Proteases and protein degradation in Escherichia coli . Experientia. 1992;48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- R48.Mayr J., Lupas A., Kellermann J., Eckerskorn C., Baumeister W., Peters J. A hyperthermostable protease of the subtilisin family bound to the surface layer of the archaeon Staphylothermus marinus . Curr. Biol. 1996;6:739–749. doi: 10.1016/s0960-9822(09)00455-2. [DOI] [PubMed] [Google Scholar]
- R49.Morikawa M., Izawa Y., Rashid N., Hoaki T., Imanaka T. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp . Appl. Environ. Microbiol. 1994;60:4559–4566. doi: 10.1128/aem.60.12.4559-4566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R50.Nelson K.E., Clayton R.A., Gill S.R., et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima . Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- R51.Porankiewicz J., Wang J., Clarke A.K. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 1999;32:449–458. doi: 10.1046/j.1365-2958.1999.01357.x. [DOI] [PubMed] [Google Scholar]
- R52.Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. J. Biol. Chem. 1993;268:6065–6068. [PubMed] [Google Scholar]
- R53.Ruepp A., Eckerskorn C., Bogyo M., Baumeister W. Proteasome function is dispensable under normal but not under heat shock conditions in Thermoplasma acidophilum . FEBS Lett. 1998;425:87–90. doi: 10.1016/s0014-5793(98)00205-1. [DOI] [PubMed] [Google Scholar]
- R54.Sako Y., Croocker P.C., Ishida Y. An extremely heat-stable extracellular proteinase (aeropyrolysin) from the hyperthermophilic archaeon Aeropyrum pernix K1. FEBS Lett. 1997;415:329–334. doi: 10.1016/s0014-5793(97)01153-8. [DOI] [PubMed] [Google Scholar]
- R55.Schmidt M., Lupas A.N., Finley D. Structure and mechanism of ATP-dependent proteases. Curr. Opin. Chem. Biol. 1999;3:584–591. doi: 10.1016/s1367-5931(99)00013-7. [DOI] [PubMed] [Google Scholar]
- R56.Schumann W. FtsH—a single-chain charonin? FEMS Microbiol. Rev. 1999;23:1–11. doi: 10.1111/j.1574-6976.1999.tb00389.x. [DOI] [PubMed] [Google Scholar]
- R57.Schäfer T., Selig M., Schönheit P. Acetyl-CoA synthetase (ADP forming) in archaea, a novel enzyme involved in acetate formation and ATP synthesis. Arch. Microbiol. 1993;159:72–83. [Google Scholar]
- R58.Schönheit P., Schäfer T. Metabolism of hyperthermophiles. World J. Microbiol. Biotechnol. 1995;11:26–57. doi: 10.1007/BF00339135. [DOI] [PubMed] [Google Scholar]
- R59.She Q., Singh R.K., Confalonieri F., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R60.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R61.Tamura N., Lottspeich F., Baumeister W., Tamura T. The role of tricorn protease and its aminopeptidase-interacting factors in cellular protein degradation. Cell. 1998;95:637–648. doi: 10.1016/s0092-8674(00)81634-7. [DOI] [PubMed] [Google Scholar]
- R62.Tomoyasu T., Mogk A., Langen H., Goloubinoff P., Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Eschirichia coli cytosol. Mol. Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- R63.Valdes-Stauber N., Scherer S. Nucleotide sequence and taxonomical distribution of the bacteriocin gene lin cloned from Brevibacterium linens M18. Appl. Environ. Microbiol. 1996;62:1283–1286. doi: 10.1128/aem.62.4.1283-1286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R64.Voorhorst W.G., Eggen R.I., Geerling A.C., Platteeuw C., Siezen R.J., de Vos W.M. Isolation and characterization of the hyperthermostable serine protease, pyrolysin, and its gene from the hyperthermophilic archaeon Pyrococcus furiosus . J. Biol. Chem. 1996;271:20426–20431. doi: 10.1074/jbc.271.34.20426. [DOI] [PubMed] [Google Scholar]
- R65.Voorhorst W.G., Warner A., de Vos W.M., Siezen R.J. Homology modelling of two subtilisin-like proteases from the hyperthermophilic archaea Pyrococcus furiosus and Thermococcus stetteri . Protein Eng. 1997;10:905–914. doi: 10.1093/protein/10.8.905. [DOI] [PubMed] [Google Scholar]
- R66.Wilson H.L., Aldrich H.C., Maupin-Furlow J. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: purification, characterization, and gene sequence analysis. J. Bacteriol. 1999;181:5814–5824. doi: 10.1128/jb.181.18.5814-5824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R67.Wilson H.L., Ou M.S., Aldrich H.C., Maupin-Furlow J. Biochemical and physical properties of the Methanococcus jannaschii 20S proteasome and PAN, a homolog of the ATPase (Rpt) subunits of the eucaryal 26S proteasome. J. Bacteriol. 2000;182:1680–1692. doi: 10.1128/jb.182.6.1680-1692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R68.Zillig W., Stetter K.O., Wunderl S., Schulz W., Priess H., Scholz I. The Sulfolobus “Caldariella” group: taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Arch. Microbiol. 1980;125:259–269. [Google Scholar]