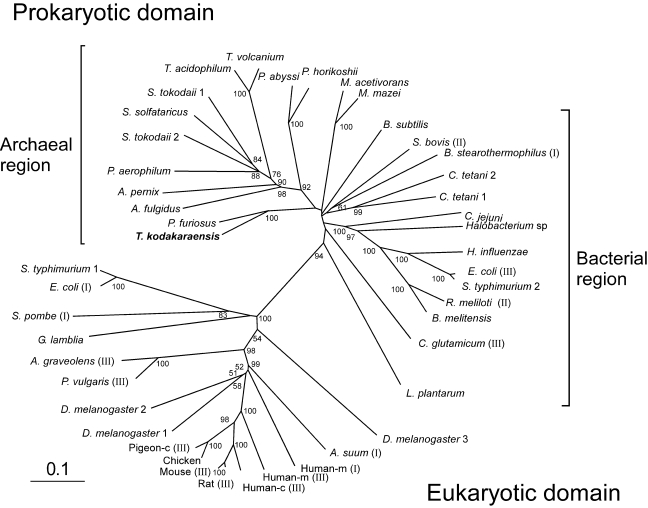
Figure 2.
Radial phylogenetic tree of representative bacterial, eukaryotic and archaeal malic enzymes. Multiple sequence alignments and phylogenetic analysis were performed with the neighbor-joining method using the the ClustalW program provided by the DNA Data Bank of Japan (DDBJ). Scale bar indicates 10 substitutions per 100 amino acids. Bootstrap resampling was performed 1000 times and only values observed in more than 50% of the replicas are shown. The accession numbers of the sequences are as follows: Aeropyrum pernix, G72731; Apium graveolens (III), Q9SDL2; Archaeoglobus fulgidus, F69465; Ascaris suum (III), S29742; Bacillus stearothermophilus (I), P16468; Bacillus subtilis, O34962; Brucella melitensis, AI3372; Campylobacter jejuni, Q9PN12; chicken, AF408407; Clostridium tetani1, AE015937; C. tetani 2, AE015944; Corynebacterium glutamicum (III), CAF18947; Drosophila melanogaster 1, Q9U1J0; D. melanogaster 2, AAF43603; D. melanogaster 3, Q9U1J2; Escherichia coli (I), P26616; E. coli (III), P76558; Giardia lamblia, U59300; Haemophilus influenzae, P43837; Halobacterium sp. NRC-1, A84315;human cytosolic (Human-c) (III), P48163; human mitochondrial (Human-m) (I), P23368; Human-m (III), Q16798; Lactobacillus plantarum, Q88XT0; Methanosarcina acetivorans, AAM05142; Methanosarcina mazei, AAM32328; mouse (III), P06801; Phaseolus vulgaris (III), P12628; pigeon cytosolic (Pigeon-c) (III), P40927; Pyrobaculum aerophilum, AAL63657; Pyrococcus abyssi, G75131; Pyrococcus furiosus, AAL81150; Pyrococcus horikoshii, H71072; rat (III), P13697; Rhizobium meliloti (II), O30807; Salmonella typhimurium 1, AE008768; S. typhimurium 2, Q9ZFV8; Schizosaccharomyces pombe (I), P40375; Streptococcus bovis (II), U35659; Sulfolobus solfataricus, A90465; Sulfolobus tokodaii1, BAB65071; S. tokodaii2, BAB65246; Thermococcus kodakaraensis,AB195292;Thermoplasma acidophilum, Q9HKY7; Thermoplasma volcanium, BAB59923. Roman numerals in parentheses indicate the class of each malic enzyme previously characterized: I, EC 1.1.1.38; II, EC 1.1.1.39; and III, EC 1.1.1.40.