Abstract
The physiological role of the nonspecific DNA-binding protein Sso7d from the crenarchaeon Sulfolobus solfataricus is unknown. In vitro studies have shown that Sso7d promotes annealing of complementary DNA strands (Guagliardi et al. 1997), induces negative supercoiling (Lopez-Garcia et al. 1998), and chaperones the disassembly and renaturation of protein aggregates in an ATP hydrolysis-dependent manner (Guagliardi et al. 2000). In this study, we examined the relationships among the binding of Sso7d to double-stranded DNA, its interaction with protein aggregates, and its ATPase activity. Experiments with 1-anilinonaphthalene-8-sulfonic acid as probe demonstrated that exposed hydrophobic surfaces in Sso7d are responsible for interactions with protein aggregates and double-stranded DNA, whereas the site of ATPase activity has a non-hydrophobic character. The interactions of Sso7d with double-stranded DNA and with protein aggregates are mutually exclusive events, suggesting that the disassembly activity and the DNA-related activities of Sso7d may be competitive in vivo. In contrast, the hydrolysis of ATP by Sso7d is independent of the binding of Sso7d to double-stranded DNA or protein aggregates.
Keywords: ATP hydrolysis, DNA-binding protein, hydrophobic protein interaction, protein aggregation
Introduction
Members of the kingdom Euryarchaeota, in the domain Archaea, have chromosomal proteins that resemble eukaryotic histones in both structure and function (Sandman and Reeve 2000). In contrast, the hyperthermophilic crenarchaea of the order Sulfolobalesexpress large amounts of basic, monomethylated, nonspecific DNA-binding proteins of about 7 kDa, which have high sequence identity among them and lack obvious similarity to any other known protein. An understanding of the physiological role of these proteins has been hampered by a lack of knowledge about many DNA-related events in crenarchaea, as well as the lack of molecular tools with which to obtain targeted mutants in Sulfolobus species.
The 7-kDa proteins from Sulfolobus spp. consist of a five-stranded, incomplete β-barrel capped at the opening by a C-terminal α-helix; they bind to the minor groove of a DNA duplex via the triple-stranded β-sheet (Agback et al. 1998, Gao et al. 1998, Robinson et al. 1998, Edmondson and Shriver 2001). The topology of the Sulfolobus 7-kDa proteins was found to be similar to that of chromatin organization modifier (chromo) domains (Ball et al. 1997) and eukaryotic SH3 domains (Knapp et al. 1998), which are involved in protein–protein interactions. Two 7-kDa DNA-binding proteins—Sso7d and Sso7c—have been identified in S. solfataricus (Oppermann et al. 1998). In vitro studies have shown that Sso7d promotes the annealing of complementary DNA strands (Guagliardi et al. 1997), induces negative supercoiling (Lopez-Garcia et al. 1998), and chaperones the disassembly and renaturation of protein aggregates in an ATP hydrolysis-dependent manner (Guagliardi et al. 2000).
The discovery of different activities in a DNA-binding protein as small as Sso7d is amazing. The available structural information is insufficient for conjecture about the locations of the ATPase site or the protein-binding surface. In this study, we focused on the relationship among the binding of Sso7d to double-stranded (ds) DNA, its interaction with protein aggregates and its ATPase activity, with the objective of determining the sites involved in each of these activities. Experiments with the probe 1-anilinonaphthalene-8-sulfonic acid (ANS) showed that Sso7d has exposed hydrophobic surfaces that interact with ds DNA and protein aggregates, whereas a non-hydrophobic surface accommodates the ATPase site. The interactions of Sso7d with ds DNA and protein aggregates are mutually exclusive events, whereas the hydrolysis of ATP is independent of the binding of Sso7d to ds DNA or to protein aggregates. The significance of these results in relation to the physiological role of Sso7d is discussed.
Materials and methods
The Sso7d was purified from the cytosol of S. solfataricus as previously described (Guagliardi et al. 2000). Lysozyme from chicken egg white (14.4 kDa; 183 U mg–1) and melittin (2.8 kDa) were from Sigma. 1-Anilinonaphthalene-8-sulfonic acid (ANS) was from Aldrich and [γ-32P]ATP (1.11 × 1014 Bq mmol–1) was from Amersham. The DNA oligomers were from Primm (Milan, Italy). All other chemicals were reagent grade.
Protein concentrations were determined by the Bradford assay (Bradford 1976) using bovine serum albumin as the standard. The Sso7d concentration was determined based on a millimolar extinction coefficient of 8.4 at 280 nm. The stock solution of ANS was prepared in water, and the concentration was determined by absorbance at 350 nm based on a millimolar extinction coefficient of 5. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 15% acrylamide) was carried out according to Laemmli (Laemmli 1970). All DNA manipulations were performed according to standard techniques (Sambrook et al. 1989).
Fluorescence measurements
Fluorescence measurements were made with a Perkin-Elmer spectrofluorimeter (Model LS 50B) at 25 °C. The excitation and emission slit width were set to 10 nm, and 0.3-ml cuvettes with a 1-cm path length were used. For tryptophan fluorescence experiments, an excitation wavelength of 295 nm was used, and the emission was recorded between 310 and 550 nm. Quenching of tryptophan fluorescence after addition of ANS was analyzed according to the modified plot of Stern-Volmer (Eftink and Ghiron 1981): the F0/(F0 – F) ratio—where F0 and F are the fluorescence intensities at 347 nm in the absence and presence of ANS, respectively—plotted against the reciprocal of ANS concentration yields a straight line whose intercept on the x-axis equals the value of the dissociation constant (Kd) for ANS. Titrations with ANS were performed manually by the addition of small volumes of a concentrated ANS solution to solutions of Sso7d in 50 mM Tris-HCl pH 7.5 at room temperature; samples were excited at 350 nm, and the emission was recorded between 400 and 600 nm. The intercept on the x-axis in a double-reciprocal plot of the fluorescence intensities at 460 nm (corrected for the absorption of the probe) versus the ANS concentration gave the value of Kd for ANS.
Band-shift assay
The double-stranded oligomer TCGACTGCGGTCAGTTAGTCACTTCCG was used as a probe. The standard mixture for the band-shift assay contained the 32P-labeled DNA probe (about 2 × 104 cpm) and 1 µg of protein in 50 mM sodium phosphate pH 8.0, 25 mM KCl, 25 mM NaCl (15 µl final volume). After incubation for 10 min at 50 °C, 3 µl of loading solution (0.25% bromophenol blue, 0.25% xylene cyanol and 30% glycerol) was added, and the sample was electrophoresed at room temperature on a native 5% polyacrylamide gel in 0.5× Tris-borate buffer at a constant voltage (10 V cm–1). After electrophoresis, the gel was dried and exposed to X-ray film.
Preparation of lysozyme aggregates
A solution of 10 mM Tris-HCl pH 7.5 containing 0.2 mg ml–1 lysozyme was incubated at 80 °C for 1.5 h, and the precipitate was pelleted by centrifugation at 20,000 g for 20 min. Molecular sieving chromatography of the suspended pellets on a Superose-6 column (1 × 70 cm, Pharmacia Biotech, Amersham Biosciences; eluent: 10 mM Tris-HCl pH 7.5, 0.2 M NaCl; flow rate 12 ml h–1) yielded aggregates in the range of 150–500 kDa.
Preparation of the ANS–Sso7d complex
The ANS–Sso7d complex was freshly prepared as follows. Ten µM Sso7d was incubated for 1 h at 25 °C with 100 µM ANS in 50 mM Tris-HCl pH 7.5. Excess ANS was removed by gel filtration chromatography on a HiLoad Superdex-75 column (1 × 30 cm, Pharmacia Biotech, Amersham Biosciences; eluent: 10 mM Tris-HCl pH 8.4, 0.2 M NaCl; flow rate 18 ml h–1). The complex recovered from the column was concentrated by vacuum centrifugation.
Preparation of the melittin–Sso7d complex
Melittin was purified by gel filtration chromatography on a BioSep-SEC-S2000 column (0.78 × 30 cm, Phenomenex, Torrance, CA; eluent: 0.1 M sodium phosphate pH 7.0, 0.1 M NaCl; flow rate 48 ml h–1) connected to a high performance liquid chromatography (HPLC) system. The melittin–Sso7d complex was freshly prepared as follows. Thirty-five µM Sso7d was incubated for 30 min at 50 °C with an equimolar amount of pure melittin in 10 mM Tris-HCl pH 7.5, and the solution was loaded onto a HiLoad Superdex-75 column (1 × 30 cm; eluent: 10 mM Tris-HCl pH 8.4, 0.2 M NaCl; flow rate 18 ml h–1). The complex recovered from the column was concentrated by vacuum centrifugation.
Preparation of the DNA–Sso7d complex
Six µM Sso7d was incubated for 10 min at 50 °C with an equimolar amount of ds GCCATGGC oligomer in 10 mM Tris-HCl pH 7.5, and then loaded onto a HiLoad Superdex-75 column (1 × 30 cm; eluent: 10 mM Tris-HCl pH 8.4, 0.2 M NaCl; flow rate 18 ml h–1). The complex recovered from the column was concentrated by vacuum centrifugation.
Enzymatic assays
Lysozyme activity was assayed at 25 °C in 1 ml of a fresh suspension of 0.1 mg ml–1 of lyophilized Escherichia coli cells in 50 mM Tris-HCl pH 7.4; 1 U min–1 was required for a decrease in absorbance of 0.1 at 350 nm. The ATPase activity of Sso7d was assayed under standard conditions with 10 µg of protein and 5.6 × 105 Bq of [γ-32P]ATP in 150 µl of 50 mM sodium phosphate pH 7.5, 2 mM ATP, 5 mM MgCl2. After incubation for 5 min at 70 °C, a 25-µl aliquot was drawn from the assay mixture, added to 0.5 ml of a suspension containing 50 mM HCl, 5 mM H3PO4 and 7% activated charcoal, and centrifuged at 4,000 g for 20 min. The radioactivity of the supernatant was determined from a 100-µl aliquot. For rate calculations, corrections were made for the amount of spontaneous ATP hydrolysis in the absence of Sso7d.
Results
Exposed hydrophobic surfaces in Sso7d are responsible for binding to ds DNA and interaction with protein aggregates
The Sso7d was purified from the cytosol of S. solfataricus by a chromatographic procedure that exploits its small size (Guagliardi et al. 2000). The purification protocol yields DNA-free Sso7d (confirmed by ethidium bromide staining of agarose gels (data not shown)) without inactivating its ATPase activity.
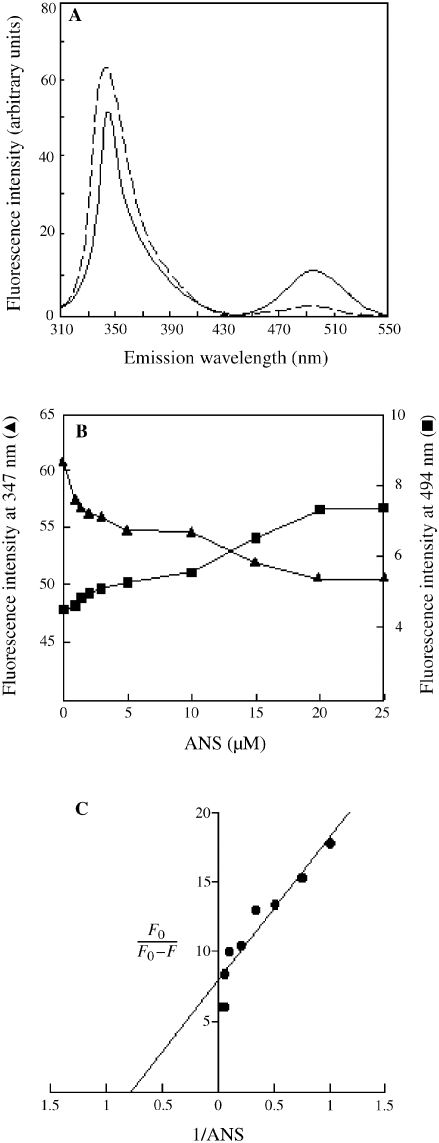
A single tryptophan residue, Trp23, is located on the surface of Sso7d; its side chain plays a major role in the nucleic acid binding process (Agback et al. 1998, Gao et al. 1998). The fluorescent probe ANS binds to hydrophobic protein surfaces (Cardamone and Puri 1992). When ANS binds close to a tryptophan residue, the intrinsic fluorescence of the protein is quenched as a result of an energy transfer from the amino acid to the probe (Cantor and Schimmel 1980). We found that the tryptophan fluorescence at 347 nm of a solution of Sso7d excited at 295 nm was quenched in the presence of ANS, concurrent with an increase in fluorescence at 494 nm (Figure 1A). Figure 1B shows the dependence of the energy transfer between tryptophan and the probe on the concentration of ANS; a Kd value of 1.29 µM was obtained for the binding of ANS when the tryptophan quenching data were analyzed by the modified plot of Stern-Volmer (Figure 1C). The experiment demonstrates that ANS binds to Sso7d at the DNA-binding surface.
Figure 1.
Binding of 1-anilinonaphthalene-8-sulfonic acid (ANS) at the Sso7d DNA-binding surface. Details of the experiment are provided in Materials and methods. (A) Tryptophan fluorescence emission spectra of 3 µM Sso7d with (broken line) and without (solid line) 25 µM ANS. (B) Dependence on ANS concentration of the energy transfer from tryptophan 23 of Sso7d to the probe. Tryptophan fluorescence emission spectra of samples containing 3 µM Sso7d and ANS at the concentrations indicated were recorded as in (A), and the maximal emission intensities (in arbitrary units) at 347 and 494 nm were plotted against ANS concentration. (C) Modified Stern-Volmer plot of tryptophan quenching data.
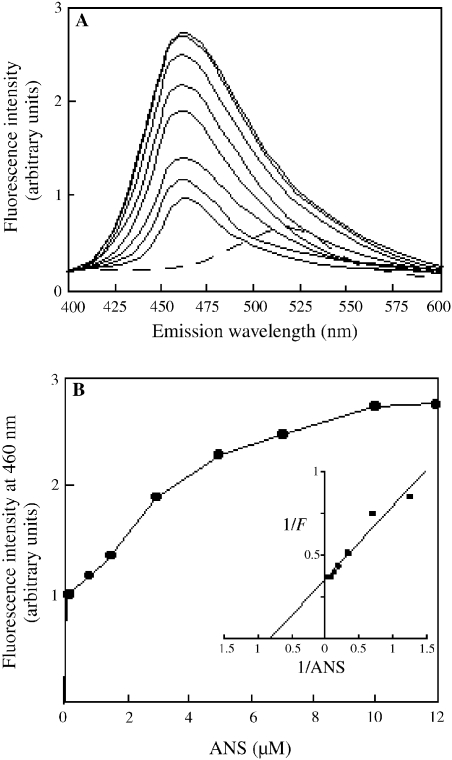
A titration experiment with Sso7d and ANS was performed to determine the binding stoichiometry between the protein and the probe. As observed when ANS binds any protein (Cardamone and Puri 1992), the fluorescence emission of ANS in the presence of Sso7d underwent a blue shift and was enhanced in a probe concentration-dependent manner (Figure 2A). The titration plot showed saturation of ANS-binding sites in Sso7d at a protein to probe molar ratio of 1:5 (Figure 2B). A double-reciprocal plot of the titration data gave a Kd value of 1.25 µM for the binding of ANS to Sso7d (Figure 2B, inset), which is not significantly different than the Kd value determined by tryptophan quenching data. Therefore, we prepared an ANS–Sso7d complex in which the exposed hydrophobic surfaces of Sso7d were shielded by the probe (see Materials and methods), and ascertained that this complex was stable for more than 30 min at 80 °C. We investigated the ability of the ANS–Sso7d complex to bind DNA and to interact with protein aggregates.
Figure 2.
Titration of Sso7d with 1-anilinonaphthalene-8-sulfonic acid (ANS). Details are provided in the Materials and methods. (A) Fluorescence emission spectra of 0.1, 0.8, 1.5, 3, 5, 7, 10 and 12 µM ANS (solid lines, from bottom to top) in the presence of 2 µM Sso7d; the spectrum of ANS alone is also reported (broken line). (B) Fluorescence titration curve of Sso7d with ANS; the inset shows the double-reciprocal plot of fluorescence titration data.
In vitro, Sso7d binds DNA without sequence specificity, and has a preference for ds DNA (Baumann et al. 1994). We observed binding of Sso7d to a ds DNA oligomer in a gel retardation experiment (Figure 3, Lane 2). The ANS–Sso7d complex, however, did not bind the DNA oligomer under the same experimental conditions (Figure 3, Lane 3).
Figure 3.
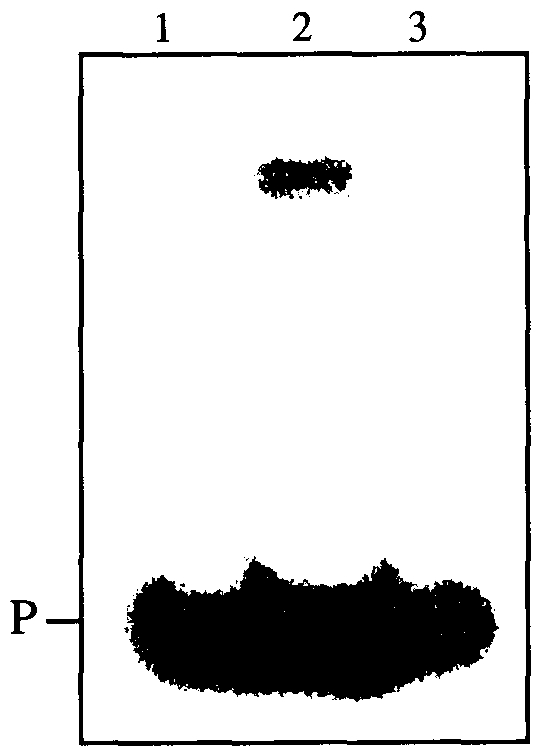
The DNA-binding activities of Sso7d and the ANS–Sso7d complex.The ANS–Sso7d complex was prepared as described in Materials and methods. The Sso7d (Lane 2) and the ANS–Sso7d complex (Lane 3) (1.5 µg protein per lane) were utilized for the band-shift assay performed as described in Materials and methods. Lane 1 contains the double-stranded DNA probe only (P).
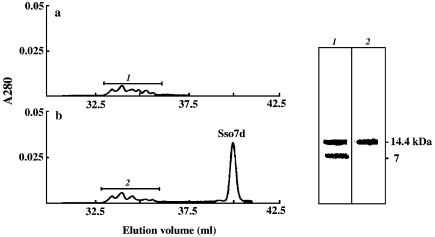
We have previously shown that Sso7d interacts with inactive aggregates of S. solfataricus γ-glycosidase in the absence of nucleotides, but that the renaturation of γ-glycosidase requires the hydrolysis of ATP by Sso7d (Guagliardi et al. 2000). In this study, we prepared inactive aggregates of lysozyme and, by means of gel filtration chromatography, examined the abilities of Sso7d and the ANS–Sso7d complex to interact with these aggregates (see Figure 4). The aggregates were incubated with Sso7d in the absence of nucleotide, and the mixture was loaded onto a gel filtration column. Spectrophotometric analysis of the eluate revealed the presence of inactive aggregates and no free Sso7d (Figure 4a); SDS-PAGE analysis showed that the Sso7d had co-eluted with the aggregates (Figure 4, right panel, Lane 1). A gel filtration column loaded with lysozyme aggregates that had been incubated with the ANS– Sso7d complex separated the inactive aggregates from the complex (Figure 4b); SDS-PAGE analysis confirmed that no Sso7d molecules were present in the fractions containing the aggregates (Figure 4, right panel, Lane 2).
Figure 4.
Interactions of Sso7d and the ANS–Sso7d complex with lysozyme aggregates. Left: profile of eluate from Superose-6 gel filtration columns loaded with 50 µg of lysozyme aggregates (prepared as described in Materials and methods) that had been incubated for 20 min at 50 °C with (a) 20 µg of Sso7d or (b) 20 µg of the ANS–Sso7d complex in 0.5 ml of 10 mM Tris-HCl pH 7.5. Columns were eluted with 10 mM Tris-HCl pH 7.5, 0.2 M NaCl at a flow rate of 12 ml h–1. Lysozyme activity was assayed as described in Materials and methods. Right: silver-stained SDS-PAGE gel. Lane 1: 5-µg sample from column (a). Lane 2: 5-µg sample from column (b).
Together, these experiments show that exposed hydrophobic surfaces in Sso7d are responsible for both the binding to ds DNA and the interaction with protein aggregates.
The interactions of Sso7d with ds DNA and with protein aggregates are mutually exclusive events
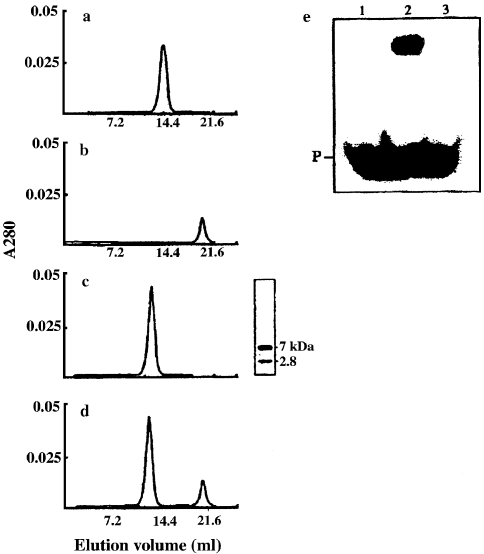
Next, we investigated whether the same or different exposed hydrophobic regions are involved in the interactions with DNA and with protein aggregates. Preliminary results showed that binding of Sso7d to lysozyme aggregates subsequently abolished binding of Sso7d to DNA (data not shown). We searched for a protein substrate whose binding to Sso7d would cause less steric hindrance than binding of the protein aggregates. Earlier studies in our laboratory showed that Sso7d binds to denatured proteins, but not to native proteins, indicating that Sso7d (like all bona fide chaperones) interacts with hydrophobic protein patches. Therefore, we tested the ability of Sso7d to interact with melittin, a hydrophobic 26-amino acid polypeptide from bee venom (Figure 5). Equimolar amounts of Sso7d and melittin were incubated for 30 min at 50 °C, and the mixture was loaded onto a gel filtration column. The column profile (Figure 5c) contained one peak that, based on SDS-PAGE analysis, contained the melittin–Sso7d complex. When Sso7d was incubated with a twofold molar excess of melittin under the same conditions, the column profile (Figure 5d) showed peaks representing free melittin and the melittin–Sso7d complex. Together, these results show binding of Sso7d to melittin, and suggest a binding stoichiometry of 1:1. Incubation of the melittin–Sso7d complex with protein aggregates did not result in disassembly of the aggregates and restoration of enzyme activity (data not shown), demonstrating that melittin binds to Sso7d at the protein aggregate-binding surface. A melittin–Sso7d complex was thus prepared (see Materials and methods) and tested for DNA-binding activity (Figure 5e). It was found that the melittin–Sso7d complex did not bind to ds DNA (Figure 5e, Lane 3). This result confirms that Sso7d lacks DNA-binding activity when it is bound to protein aggregates.
Figure 5.
Interaction of Sso7d with melittin, and DNA-binding activity of the melittin–Sso7d complex. Superdex-75 gel filtration columns loaded with 0.5 ml of 10 mM Tris-HCl pH 7.5 containing: (a) 35 µM Sso7d; (b) 35 µM melittin; (c) 35 µM Sso7d and 35 µM melittin; or (d) 35 µM Sso7d and 70 µM melittin after incubation at 50 °C for 30 min. Columns were eluted with 10 mM Tris-HCl pH 7.5, 0.2 M NaCl at a flow rate of 18 ml h–1. The SDS-PAGE analysis of the peak from column (c) (5 µg) is reported; after electrophoresis, the gel was silver-stained. (e) Sso7d (Lane 2) and the melittin–Sso7d complex (Lane 3) (1.5 µg of protein per lane) were utilized for the band-shift assay performed as described in Materials and methods; Lane 1 contains the double-stranded DNA probe only (P). The melittin–Sso7d complex was prepared as described in Materials and methods.
X-Ray crystallography has shown that a double-stranded eight-nucleotide oligomer covers the DNA-binding surface of Sso7d (Gao et al. 1998). With the experimental strategy depicted in Figure 4, we obtained evidence that Sso7d, in complex with the ds oligomer GCCATGGC (see Materials and methods), did not interact with the protein aggregates.
These results show that the interactions of Sso7d with ds DNA and with protein aggregates are mutually exclusive events, and thus, that ds DNA and protein aggregates bind to the same or overlapping hydrophobic surfaces of Sso7d.
ATP hydrolysis is independent of the binding of Sso7d to DNA or to protein aggregates
The intrinsic ATPase activity of Sso7d has been biochemically characterized (Guagliardi et al. 2000), but the protein contains no known ATPase domains. The ANS–Sso7d complex and Sso7d had the same Km value for the nucleotide and the same maximal velocity for the hydrolysis of ATP (Table 1); that is, the binding of ANS to Sso7d did not affect the binding or hydrolysis of the nucleotide. It is possible that the ATPase site is located in a non-hydrophobic region of the protein.
Table 1.
Effects of the binding of Sso7d to melittin, DNA or ANS on its ATPase activity. The melittin–Sso7d, DNA–Sso7d and ANS– Sso7d complexes were prepared as described in Materials and methods. The ATPase activity was assayed as described in Materials and methods. Each assay was performed in duplicate. The Km value was calculated from a double-reciprocal plot of initial velocities versus ATP concentrations in the range 0.0 5–1 mM.
| Vmax (pmol Pi min–1 µg–1) | Km (mM) | |
| Sso7d | 13.6 | 0.2 |
| Melittin–Sso7d complex | 13.2 | 0.2 |
| DNA–Sso7d complex | 13.8 | 0.2 |
| ANS–Sso7d complex | 13.4 | 0.2 |
The complex between Sso7d and protein aggregates disassembles into native protein molecules upon the addition of hydrolyzable ATP (Guagliardi et al. 2000), indicating that Sso7d carries out the hydrolysis of the nucleotide while bound to the protein aggregates. However, characterization of the ATPase activity of Sso7d bound to protein aggregates is prevented by the rapid dissociation of the complex under conditions of nucleotide hydrolysis. We found that both the melittin–Sso7d complex and the ds DNA–Sso7d complex had an associated ATPase activity with a maximal hydrolysis rate and a Km value for ATP that were comparable to values for Sso7d (Table 1). It is evident that binding of Sso7d to the protein aggregates or to ds DNA does not interfere with the hydrolysis of ATP, suggesting that the ATPase site is independent of the ds DNA- and protein aggregate-binding sites.
Discussion
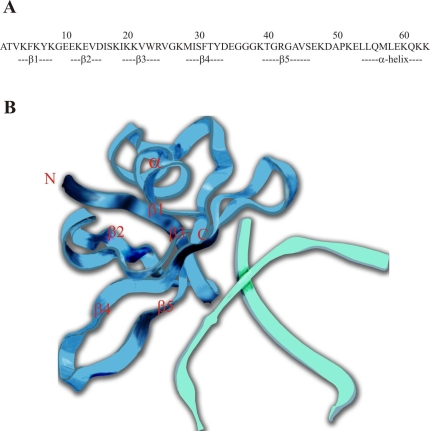
The protein Sso7d is a monomeric, globular protein with a small hydrophobic core and many solvent-exposed hydrophobic amino acids (Agback et al. 1998, Gao et al. 1998). The crystal structure of Sso7d in complex with a double-stranded DNA octamer (Gao et al. 1998) revealed that the triple-stranded β-sheet (β3, β4 and β5 strands) constitutes a large DNA-binding surface, where several exposed hydrophobic amino acids (including Trp23) are involved in binding Sso7d to the minor groove of the DNA (Figure 6). In agreement with these findings, our results show that the binding of ANS to Sso7d leads to tryptophan fluorescence quenching and abolition of the DNA-binding activity of Sso7d.
Figure 6.
(A) Sequence and secondary structure elements of Sso7d. (B) View of Sso7d in complex with a double-stranded DNA octamer (from NDB Atlas).
When complexed with ANS, Sso7d does not bind protein aggregates, indicating that hydrophobic surfaces of Sso7d are necessary for interaction with the aggregates. The discovery that binding of a ds DNA octamer to Sso7d prevents the subsequent interaction of Sso7d with protein aggregates, and vice versa, suggests that the hydrophobic surfaces of Sso7d involved in binding protein aggregates and ds DNA are the same, or that they overlap. Ball et al. (1997), who recognized the structural similarity between Sso7d and the chromatin organization modifier (chromo) domain that functions as a protein interaction motif in several proteins involved in chromatin structure formation and transcriptional control, have suggested that the site of protein binding in Sso7d may be the hydrophobic groove formed by the β3, β4 and β5 strands.
There are no known ATPase domains in Sso7d. We showed that Sso7d in complex with melittin or ds DNA displays ATPase activity. Moreover, we found that tryptophan fluorescence was not quenched following the binding of ATP to Sso7d (data not shown). Together, these results show that the ATPase site is independent of the binding surfaces for protein aggregates or ds DNA. Based on the experiments with ANS, the ATPase site has a non-hydrophobic character. The charged amino acids of Sso7d are not uniformly distributed on the surface (Gao et al. 1998). The β1 and β2 strands and the C-terminal segment of Sso7d do not interact with DNA, and contain many glutamic acid, aspartic acid and lysine residues. Such residues are essential for the activity of several canonical ATPases. Mutational studies will define the residues responsible for the ATPase activity of Sso7d.
Evidence that binding of Sso7d to DNA precludes binding to protein aggregates sustains the possibility that Sso7d may assist different events of DNA metabolism. The involvement of Escherichia coli RecA (probably the best studied ATP-dependent multifunctional DNA-binding protein) in both genetic recombination and DNA repair is regulated by the overlap of the binding surface for DNA with the binding surface for a repressor protein (Takahshi et al. 1996). The ability of Sso7d to promote annealing of complementary DNA strands and to induce negative supercoiling suggests that it could protect DNA from denaturation by high temperatures. In Bacteria and Eukarya, initiation of replication requires the timely disassembly and reactivation of inactive multimeric factors by the ATP-dependent chaperones of the heat-shock family hsp70; however, counterparts of these chaperones have not been found in hyperthermophiles (Macario and Conway de Macario 2001). Among the proteins present in crude extracts from S. solfataricus, only Sso7d, which is not a heat-shock protein of the archaeon, disassembles protein aggregates in vitro (Guagliardi et al. 2000), whereas the heat-shock protein called chaperonin (like all prokaryotic and eukaryotic chaperonins) promotes refolding of denatured proteins by preventing aggregation among the intermediates, but does not dissolve aggregates once they form (Guagliardi et al. 1995). It is tempting to speculate that Sso7d fulfils, in the archaeon, the functions of hsp70 chaperones. The fact that Sso7d represents one of the most abundant proteins of S. solfataricus (about 4% of the total cytosolic protein content) supports the hypothesis that it could promote events that require protein factors in order to occur at high temperatures, such as the renaturation of DNA and the disaggregation of aggregated proteins.
Acknowledgments
This work was supported by the EU Program “Extremophiles as Cell Factories,” by the Target Project on Biotechnology of the CNR, and by MURST (PRIN 1999 and 2000).
References
- R1.Agback P.H., Baumann S., Knapp R., Ladenstein R., Hard T. Architecture of nonspecific protein–DNA interactions in the Sso7d–DNA complex. Nat. Struct. Biol. 1998;5:579–584. doi: 10.1038/836. [DOI] [PubMed] [Google Scholar]
- R2.Ball L.J., Murzina N.V., Broadhurst R.W., et al. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 1997;16:2473–2481. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Baumann H., Knapp S., Lundback T., Ladenstein R., Hard T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus . Nat. Struct. Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- R4.Bradford M.M. A rapid and sensitive method for quantitation of microgram quantities of proteins utilizing dye binding of proteins. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- R5.Cantor C.R., Schimmel P.R. San Francisco: Part III: the behavior of biological macromolecules. W.H. Freeman; 1980. Biophysical chemistry; pp. 433–455. [Google Scholar]
- R6.Cardamone M., Puri N.K. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem. J. 1992;282:589–593. doi: 10.1042/bj2820589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Edmondson S.P., Shriver J.W. DNA binding proteins Sac7d and Sso7d from Sulfolobus . Methods Enzymol. 2001;334:129–145. doi: 10.1016/s0076-6879(01)34463-4. [DOI] [PubMed] [Google Scholar]
- R8.Eftink M.R., Ghiron C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981;114:199–227. doi: 10.1016/0003-2697(81)90474-7. [DOI] [PubMed] [Google Scholar]
- R9.Gao Y.G., Su S.Y., Robinson H., Padmanabhan S., Lim L., Mccrary B.S., Edmondson S.P., Shriver J.W., Wang A.H. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat. Struct. Biol. 1998;5:782–786. doi: 10.1038/1822. [DOI] [PubMed] [Google Scholar]
- R10.Guagliardi A., Cerchia L., Rossi M. Prevention of in vitro protein thermal aggregation by the Sulfolobus solfataricus chaperonin. Evidence for nonequivalent binding surfaces on the chaperonin molecule. J. Biol. Chem. 1995;270:28126–28132. doi: 10.1074/jbc.270.47.28126. [DOI] [PubMed] [Google Scholar]
- R11.Guagliardi A., Napoli A., Rossi M., Ciaramella M. Annealing of complementary DNA strands above the melting point of the duplex promoted by an archaeal protein. J. Mol. Biol. 1997;267:841–848. doi: 10.1006/jmbi.1996.0873. [DOI] [PubMed] [Google Scholar]
- R12.Guagliardi A., Cerchia L., Moracci M., Rossi M. The chromosomal protein Sso7d of the crenarchaeon Sulfolobus solfataricus rescues aggregated proteins in an ATP hydrolysis-dependent manner. J. Biol. Chem. 2000;275:31813–31818. doi: 10.1074/jbc.M002122200. [DOI] [PubMed] [Google Scholar]
- R13.Knapp S., Mattson P.T., Christova P., Berndt K.D., Karshikoff A., Vihinen M., Smith C.I., Ladenstein R. Thermal unfolding of small proteins with SH3 domain folding pattern. Proteins. 1998;31:309–319. doi: 10.1002/(sici)1097-0134(19980515)31:3<309::aid-prot7>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- R14.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- R15.Lopez-Garcia P., Knapp S., Ladenstein R., Forterre P. In vitro DNA binding of the archaeal protein Sso7d induces negative supercoiling at temperatures typical for thermophilic growth. Nucleic Acids Res. 1998;26:2322–2328. doi: 10.1093/nar/26.10.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Macario A.J.L., Conway de Macario E. The molecular chaperone system and other anti-stress mechanisms in archaea. Front. Biosci. 2001;6:262–283. doi: 10.2741/macario. [DOI] [PubMed] [Google Scholar]
- R17.Oppermann U.C.T., Knapp S., Bonetto V., Ladenstein R., Jornvall H. Isolation and structure of repressor-like proteins from the archaeon Sulfolobus solfataricus . FEBS Lett. 1998;432:141–144. doi: 10.1016/s0014-5793(98)00848-5. [DOI] [PubMed] [Google Scholar]
- R18.Robinson H., Gao Y.-G., Mccrary B.S., Edmondson S.P., Shriver J.W., Wang A.H.-J. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature. 1998;392:202–205. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- R19.Sambrook J., Fritsch E.F., Maniatis T. 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- R20.Sandman K., Reeve J.N. Structure and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch. Microbiol. 2000;173:165–169. doi: 10.1007/s002039900122. [DOI] [PubMed] [Google Scholar]
- R21.Takahshi M., Maraboeuf F., Norden B. Locations of functional domains in the RecA protein. Eur. J. Biochem. 1996;242:20–28. doi: 10.1111/j.1432-1033.1996.0020r.x. [DOI] [PubMed] [Google Scholar]